The step-5 tutorial program
| Table of contents | |
|---|---|
Introduction
In this program we compare two different implementations of the popular NIPALS algorithm for principal component analysis.
The commented program
#ifndef CUDA_KERNEL_STEP_5_CU_H #define CUDA_KERNEL_STEP_5_CU_H
Deklarationen der kernel-wrapper-Funktionen
void step_5_dummy(); #endif // CUDA_KERNEL_STEP_5_CU_H
Header-Datei der CUDA utility-Bibliothek.
#include <cutil_inline.h> #include <cuda_kernel_wrapper_step-5.cu.h>
Device Funktionen
Vor der Implementierung der Kernel werden noch ein paar Hilfsfunktionen diskutiert. Diese sogenannten "Device Funktionen" sind nur auf der GPU ausfuehrbar und automatisch inline.
Device Function: lex_index_2D
Diese Funktion berechnet aus Zeilenindex 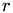 und Spaltenindex
und Spaltenindex 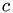 eines Matrixeintrags die Position
eines Matrixeintrags die Position 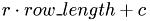 in dem linearen Array, in dem die Matrix mit der Zeilenlaenge
in dem linearen Array, in dem die Matrix mit der Zeilenlaenge 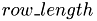 zeilenweise abgespeichert ist.
zeilenweise abgespeichert ist.
- Parameters:
-
r : Zeilenindex c : Spaltenindex row_length : Zeilenlaenge
__device__ int lex_index_2D(int r, int c, int row_length) { return c + r*row_length; }
Kernel
CUDA erweitert die Sprache C um sogenannte Kernel, deren parallele Ausfuehrung von den threads uebernommen wird. Um einen Kernel N-mal auszufuehren, muessen N threads gestartet werden. Wie das genau geht, wird spaeter an der entsprechenden Stelle erklaert.
Fuer die Parallelisierung reicht es vorerst zu wissen, dass in den Kernel-Funktionen das implementiert wird, was ein einzelner Rechenkern des Graphikchips machen soll. Allerdings sind dabei einige Feinheiten der Speicherarchitektur zu beachten, die sich auch in der Organisation der Gesamtheit aller threads niederschlaegt. Zwar kann jeder thread auf den gesamten Speicher der GPU zugreifen, allerdings dauert das einige Hundert Taktzyklen. Zur Abmilderung dieses Problems sind threads in Bloecken organisiert, innerhalb derer die threads ueber einen gemeinsamen schnellen Zwischenspeicher (shared memory) und Synchronisationspunkte kommunizieren koennen. Hingegen koennen threads aus verschiedenen Bloecken lediglich ueber den Hauptspeicher der GPU kommunizieren und besitzen keine Moeglichkeit der Synchronisation, laufen also voellig unabhaengig voneinander.
Die Besonderheit am shared memory ist, dass zwar die Zugriffszeiten vernachlaessigbar sind, dafuer aber immer 16 threads (ein sogenannter half-warp) gleichzeitig zugreifen, wodurch random access in der Regel enge Grenzen gesetzt sind. Als Beispiel, wie so ein Kernel aussehen kann
Kernel: single_thread_cholesky
Zum Aufwaermen machen wir das einfachste, was man machen kann: Wir lassen die Cholesky-Zerlegung von einem einzelnen Thread berechnen. Der Sinn davon ist, dass man den Quelltext der CPU-Implementierung einfach nur in einen CUDA-Kernel kopieren muss. Wenn das laeuft, hat man sich davon ueberzeugt, dass das Programmieren mit CUDA tatsaechlich wie mit C geht und die Programme lediglich an einem anderen Ort ausgefuehrt werden. Danach kann damit begonnen werden, die Cholesky-Zerlegung zu parallelisieren.
- Parameters:
-
A : lineares Array, in dem die Zeilen der zu faktorisierenden Matrix hintereinander weg abgespeichert sind gpu_tile_size : Laenge einer Zeile
__global__ void single_thread_cholesky(float *A, const int gpu_tile_size) { typedef float T;
Die aeussere Schleife ueber die Zeilen von L.
for (unsigned int r = 0; r < gpu_tile_size; ++r) {
Berechne den Diagonaleintrag des Cholesky-Faktors.
T sum = 0.;
unsigned int idx;
unsigned int idx_c;
Summiere die Quadrate aller bisherigen Eintraege des Cholesky-Faktors in dieser Zeile.
for (unsigned int u = 0; u < r; ++u) { idx = lex_index_2D(r, u, gpu_tile_size); sum += A[idx] * A[idx]; } idx = lex_index_2D(r, r, gpu_tile_size); A[idx] = sqrt(A[idx] - sum);
Berechne Nebendiagonal-Eintraege des Cholesky-Faktors. Dabei wird ausgenutzt, dass A symmetrisch ist. Die Hilfsvariable sum entspricht der in Schritt 2.1 des Algorithmus.
for (unsigned int c = r+1; c < gpu_tile_size; ++c) { sum = 0.; for (unsigned int u = 0; u < r; ++u) { idx_c = lex_index_2D(c, u, gpu_tile_size); idx = lex_index_2D(r, u, gpu_tile_size); sum += A[idx_c]*A[idx]; } idx_c = lex_index_2D(c, r, gpu_tile_size); idx = lex_index_2D(r, c, gpu_tile_size); A[idx_c] = A[idx] - sum; idx = lex_index_2D(r, r, gpu_tile_size); A[idx_c] /= A[idx]; } } }
Kernel: __dummy
Dieser Kernel macht nichts sinnvolles und ist nur dazu da, in der Treiber-Klasse Modell zu stehen.
__global__ void __step_5_dummy() { int i = 2; int j = 3; int k = i+j; i += k; }
Funktion: step_5_dummy
Wrapper-Funktion fuer den entsprechenden Kernel. Legt die Aufteilung der threads fest und startet diese.
void step_5_dummy() { dim3 grid(5,5); dim3 blocks(16,16); __step_5_dummy<<<grid, blocks>>>(); } #ifndef CUDADriver_STEP_5_H #define CUDADriver_STEP_5_H #include <lac/blas++.h> #include <lac/cublas_algorithms.h> #include <lac/full_matrix.h>
Jedes Beispielprogramm kommt in einen separaten naemspace, damit man die verschiedenen Treiberklassen ggf. spaeter in komplexeren Projekten kombinieren kann.
namespace step5 {
Class: CudaNIPALS
Principal component analysis by means of the NIPALS algorithm.
- Parameters:
-
blas must be one of the BLAS wrapper types.
template <typename T, typename blas> class CudaNIPALS { public: typedef typename blas_pp<T, blas>::blas_wrapper_type BW; typedef typename blas_pp<T, blas>::FullMatrixAccessor FullMatrixAccessor; typedef typename blas_pp<T, blas>::Matrix Matrix; typedef typename blas_pp<T, blas>::SubMatrix SubMatrix; typedef typename blas_pp<T, blas>::MatrixSubCol MatrixSubCol; typedef typename blas_pp<T, blas>::MatrixSubRow MatrixSubRow; typedef typename blas_pp<T, blas>::Vector Vector; CudaNIPALS(unsigned int p_n_components,unsigned int p_max_iter) : num_zero(1e-8),max_iter(p_max_iter), n_components(p_n_components) {} ~CudaNIPALS (); double get_pca (const ::FullMatrix<T>& X); double get_pca_blas_version (const ::FullMatrix<T>& X); const Matrix & scores() const { return T_d; } const Matrix & loads() const { return P_d; } const Matrix & residual() const { return R_d; } ::Vector<T> lambda; private: Matrix T_d, P_d, R_d;
run-time parameter:
unsigned int n_components; //X.n_cols();
static
const double num_zero; // = 1e-8; unsigned int max_iter;
void check_matrix_dimensions(const ::FullMatrix<T>& A);
};
Class: CudaTruncatedSVDecomposition
template <typename T, typename blas> class CudaTruncatedSVDecomposition { public: typedef typename blas_pp<T, blas>::blas_wrapper_type BW; typedef typename blas_pp<T, blas>::FullMatrixAccessor FullMatrixAccessor; typedef typename blas_pp<T, blas>::Array Array; typedef typename blas_pp<T, blas>::Matrix Matrix; typedef typename blas_pp<T, blas>::SubMatrix SubMatrix; typedef typename blas_pp<T, blas>::MatrixSubCol MatrixSubCol; typedef typename blas_pp<T, blas>::MatrixSubRow MatrixSubRow; typedef typename blas_pp<T, blas>::Vector Vector; typedef typename bw_types::algorithms<T> algorithms; CudaTruncatedSVDecomposition() {} ~CudaTruncatedSVDecomposition() {} void factorize(const ::FullMatrix<T>& A); const Matrix & V() const { return U_d; } const Matrix & U() const { return V_d; }
const Matrix & Lambda() const { return Lambda_d; }
private: Matrix R_d, U_d, V_d; ::Vector<T> __Lambda; }; } // namespace step5 END #endif // CUDADriver_STEP_5_H #ifndef CUDA_DRIVER_STEP_5_HH #define CUDA_DRIVER_STEP_5_HH #include "cuda_driver_step-5.h"
Deklarationen der kernel-wrapper-Funktionen.
#include "cuda_kernel_wrapper_step-5.cu.h" #include <cutil_inline.h> #include <base/CUDATimer.h> #include <base/timer.h>
Funktion: Konstruktor
template <typename T, typename blas> step5::CudaNIPALS<T, blas>::CudaNIPALS() {}
Funktion: Destruktor
template <typename T, typename blas> step5::CudaNIPALS<T, blas>::~CudaNIPALS() {} static const bool measure_all = true; static const bool measure_RT_times_tk = false; static const bool measure_R_times_pk = false;
Funktion: get_pca
- Parameters:
-
X,: Datenmatrix, die analysiert werden soll. Rueckgabewert ist die Nettozeit fuer die PCA.
template <typename T, typename blas> double step5::CudaNIPALS<T, blas>::CudaNIPALS::get_pca(const ::FullMatrix<T>& X) {
Leider arbeitet CUBLAS intern im column-major-Format und erwartet auch jegliche Eingabe in diesem Format. Deswegen wird die Transponierte der zu faktorisierenden Matrix in einer lokalen Kopie abgespeichert und mit dieser weitergearbeitet. Damit man auch jedes Mal wieder darueber stolpert, was man da eigentlich macht, bekommt die Kopie auch einen richtig haesslichen Namen.
::FullMatrixAccessor<T> X_t_ugly(X, true / *transpose copy* /);
Reinitialisiere die device-Darstellungen Q_d, R_d von 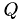 und
und 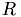 . Im Laufe der Zerlegung wird
. Im Laufe der Zerlegung wird 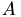 mit
mit 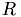 ueberschrieben, deswegen wird
ueberschrieben, deswegen wird 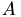 gleich in
gleich in R_d abgespeichert. Kopiere die Daten der zu faktorisierenden Matrix vom host zum device.
this->R_d = X_t_ugly;
scores
this->T_d.reinit(X.n_rows(), n_components );
loads
this->P_d.reinit(X.n_cols(), n_components );
lambda.reinit(n_components);
Ausdruck fuer die transponierte Matrix.
transpose<Matrix> R_T(this->R_d);
View auf die 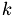 -te Spalte der load- und score-Matrix.
-te Spalte der load- und score-Matrix.
MatrixSubCol hat_t_k (this->T_d, 0, 0);
MatrixSubCol hat_p_k(this->P_d, 0, 0);
MatrixSubCol hat_r_k(this->R_d, 0, 0);
Vector p_k(hat_p_k.size() ), t_k(hat_t_k.size() );
double net_time = 0.;
CUDATimer pca_timer;
for (unsigned int k = 0; k < n_components; k++)
{
T lambda_old = 0.;
lambda(k) = 0.;
bool converged = false;
hat_t_k.reset(0, k);
hat_p_k.reset(0, k);
hat_r_k.reset(0, k);
t_k = hat_r_k;
for (unsigned int j = 0; j < max_iter //&& !converged
; j++)
{
if (measure_RT_times_tk && !measure_all)
{
pca_timer.reset();
p_k = R_T * t_k;
pca_timer.stop();
net_time += pca_timer();
}
else
p_k = R_T * t_k;
std::cout << __FUNCTION__ << " MV data : \nt_k:"; t_k.print(); std::cout << "**************** p_k :\n"; p_k.print(); R_T.A.print();
p_k /= p_k.l2_norm();
if (measure_R_times_pk && !measure_all)
{
pca_timer.reset();
t_k = R_d * p_k;
pca_timer.stop();
net_time += pca_timer();
}
else
t_k = R_d * p_k;
lambda(k) = t_k.l2_norm();
converged = (std::fabs(lambda(k) - lambda_old) <= num_zero);
lambda_old = lambda(k);
}
R_d.add_scaled_outer_product(-1., t_k, p_k);
hat_p_k = p_k;
hat_t_k = t_k;
}
for measuring overall run-time
if (measure_all && !(measure_RT_times_tk || measure_R_times_pk)) { pca_timer.stop(); net_time += pca_timer(); } return net_time; }
Funktion: get_pca_blas_version
- Parameters:
-
X,: Datenmatrix, die analysiert werden soll. Rueckgabewert ist die Nettozeit fuer die PCA.
template <typename T, typename blas> double step5::CudaNIPALS<T, blas>::CudaNIPALS::get_pca_blas_version(const ::FullMatrix<T>& X) {
Leider arbeitet CUBLAS intern im column-major-Format und erwartet auch jegliche Eingabe in diesem Format. Deswegen wird die Transponierte der zu faktorisierenden Matrix in einer lokalen Kopie abgespeichert und mit dieser weitergearbeitet. Damit man auch jedes Mal wieder darueber stolpert, was man da eigentlich macht, bekommt die Kopie auch einen richtig haesslichen Namen.
::FullMatrixAccessor<T> X_t_ugly(X, true / *Erstelle Kopie der Transponierten* /);
Reinitialisiere die device-Darstellungen Q_d, R_d von 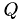 und
und 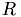 . Im Laufe der Zerlegung wird
. Im Laufe der Zerlegung wird 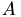 mit
mit 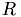 ueberschrieben, deswegen wird
ueberschrieben, deswegen wird 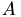 gleich in
gleich in R_d abgespeichert. Kopiere die Daten der zu faktorisierenden Matrix vom host zum device.
this->R_d = X_t_ugly;
this->P_d.reinit(n_components, X.n_cols() );
this->T_d.reinit(X.n_rows(), n_components );
this->lambda.reinit(n_components);
double net_time = 0.;
CUDATimer pca_timer;
------------- P(L)AIN BLAS -----------
int M = this->R_d.n_rows(); int N = this->R_d.n_cols(); int K = n_components; T * dR = this->R_d.array().val(); T * dT = this->T_d.array().val(); T * dP = this->P_d.array().val(); for(int k=0; k < K; k++) { BW::copy(M, &dR[k*M], 1, &dT[k*M], 1); T lambda_old = 0.; for(unsigned int j=0; j < max_iter; j++) { if (measure_RT_times_tk && !measure_all) { pca_timer.reset();
p_k = R_T * t_k;
BW::gemv('t', M, N, 1.0, dR, M, &dT[k*M], 1, 0.0, &dP[k*N], 1); pca_timer.stop(); net_time += pca_timer(); } else BW::gemv('t', M, N, 1.0, dR, M, &dT[k*M], 1, 0.0, &dP[k*N], 1); BW::scal(N, 1.0/BW::nrm2(N, &dP[k*N], 1), &dP[k*N], 1); if (measure_R_times_pk && !measure_all) { pca_timer.reset(); BW::gemv('n', M, N, 1.0, dR, M, &dP[k*N], 1, 0.0, &dT[k*M], 1); pca_timer.stop(); net_time += pca_timer(); } else BW::gemv('n', M, N, 1.0, dR, M, &dP[k*N], 1, 0.0, &dT[k*M], 1); lambda(k) = BW::nrm2(M, &dT[k*M], 1);
In Andrecut's version num_zero is still multiplied by lambda(k)
if(fabs(lambda_old - lambda(k)) < num_zero) break; lambda_old = lambda(k); } BW::ger(M, N, -1.0, &dT[k*M], 1, &dP[k*N], 1, dR, M); } if (measure_all && !(measure_RT_times_tk || measure_R_times_pk)) { pca_timer.stop(); net_time += pca_timer(); } return net_time; } template <typename T, typename blas> void step5::CudaTruncatedSVDecomposition<T, blas>::factorize(const ::FullMatrix<T>& A) { ::FullMatrixAccessor<T> A_t_ugly(A, true / *Erstelle Kopie der Transponierten* /); this->R_d = A_t_ugly;
run-time parameter
const unsigned int n_components = A.n_cols(); static const unsigned int max_iter = 1000; static const T num_zero = 1e-5; this->V_d.reinit(A.n_rows(), n_components); this->U_d.reinit(A.n_cols(), n_components); this->__Lambda.reinit (n_components); transpose<Matrix> R_T(R_d); SubMatrix V_k(V_d, 0, 0); transpose<SubMatrix> V_k_T(V_k); SubMatrix U_k(U_d, 0, 0); transpose<SubMatrix> U_k_T(U_k); Vector v_k (A.n_rows()); Vector b (A.n_rows()); Vector u_k (A.n_cols()); Vector a (A.n_cols()); int r_begin = 0; int r_end_U = U_d.n_rows(); int r_end_V = V_d.n_rows(); for (unsigned int k = 0; k < n_components; k++) { T lambda = 0., lambda_old = 0.; MatrixSubCol v_k_view(R_d, 0, k), u_k_view(U_d, 0, k), r_k(R_d, 0, k); v_k = r_k; bool converged = false;
Reorthogonalisierung
int new_c_begin = 0; int new_c_end = k; if (k>0) {
std::cout << "U_" << k << " : \n";
U_k.reset(r_begin, r_end_U, new_c_begin, new_c_end);
V_k.reset(r_begin, r_end_V, new_c_begin, new_c_end);
U_k.print();
}
for (unsigned int j = 0; j < max_iter && !converged; j++)
{
u_k = R_T * v_k;
#define USE_REORTHO
#ifdef USE_REORTHO
if (k > 0)
{
std::cout << " u_" << k << " : \n"; u_k.print();
a = U_k_T * u_k;
std::cout << "U_" << k << " * u_k : \n"; a.print();
u_k -= U_k * a;
a = U_k_T * u_k; std::cout << "after reorthogonalization : U_" << k << " * u_k : \n"; a.print();
} #endif u_k /= u_k.l2_norm(); v_k = R_d * u_k; #ifdef USE_REORTHO if (k > 0) { b = V_k_T * v_k; v_k -= V_k * b; } #endif lambda = v_k.l2_norm(); v_k /= lambda; converged = std::fabs(lambda -lambda_old) < num_zero; lambda_old = lambda;
std::cout << "iter " << j << " : lambda_" << k << " = " << std::setprecision(8) << lambda // (k) << std::endl;
}
R_d.add_scaled_outer_product(-lambda, v_k, u_k);
v_k_view = v_k;
u_k_view = u_k;
this->__Lambda(k) = lambda;
}
std::cout << "SV from tSVD : \n";
this->__Lambda.print(std::cout, 8, false);
}
#endif // CUDA_DRIVER_STEP_5_HH
STL header
#include <iostream> #include <vector> #include <QApplication> #include <QString> #include <QMessageBox> #include <QFileDialog>
Treiber fuer den GPU-Teil
#include "cuda_driver_step-5.h"
deal.II-Komponenten include <lac/vector.h>
cublas-Wrapper-Klassen. Binden alle sonstigen benoetigten header-Dateien ein.
#include "cuda_driver_step-5.hh"
Reuse GlobalData and the Params classes from step-4.
#include "../step-4/step-4.hh" namespace step5 {
Class: PCATestUIParams
struct PCATestUIParams : public step4::TestUIParamsBase { PCATestUIParams() : TestUIParamsBase() {} static void declare(::ParameterHandler & prm); void get(::ParameterHandler & prm); QStringList possible_matrices_to_print_orig; QStringList selected_orig; QStringList possible_matrices_to_print_num; QStringList selected_num; unsigned int n_components, max_iter; template<typename T> void parse_wave_forms(::FullMatrix<T> & A); private: PCATestUIParams (const PCATestUIParams & / *other* /) : TestUIParamsBase() {} PCATestUIParams & operator = (const PCATestUIParams & / *other* /) { return *this; } }; template <typename T, typename blas> class PCATest { public: typedef typename blas_pp<T, blas>::FullMatrixAccessor FullMatrixAccessor; typedef typename blas_pp<T, blas>::Matrix Matrix; typedef transpose<Matrix> tr; typedef typename blas_pp<T, blas>::SubMatrix SubMatrix; typedef typename blas_pp<T, blas>::MatrixSubCol MatrixSubCol; typedef typename blas_pp<T, blas>::Vector Vector; PCATest(::ParameterHandler & prm); void run(); private: void setup_and_assemble(unsigned int nr, unsigned int nc); void factorize(); void check_results(const ::FullMatrix<T> & A, const CudaNIPALS<T, blas>& PCAf); void save_results(); ::FullMatrix<T> Q, B, P_t, H; PCATestUIParams params; ::ConvergenceTable results_table; unsigned int n_successful_measurements; }; } void step5::PCATestUIParams::declare(::ParameterHandler &prm) { TestUIParamsBase::declare(prm);
Die Masse der Parameter dient dazu, sich die Details der Ergebnisse anschauen zu koennen.
prm.enter_subsection("Visualization flags."); prm.declare_entry ("Original matrices to print", "A", ::Patterns::MultipleSelection("A|Q|B|P_T|X|Sigma|Y_T|U|V_T"), "Possible values: A,Q,B,P_T,X,Sigma,Y_T,U,V_T " " Entries must be separated by a colon." " " "The direct SVD is a two-stage process. " "In the first factorization gives A = Q * B * P_T, " "where B is a upper bidiagonal matrix, and Q and P_T " "are orthogonal matrices. In the second factorization is " "B = X * Sigma * Y_T with S diagonal, containing the " "singular values and X and Y_T are orthogonal matrices again. " "The final result A = U * Sigma * V_T of the SVD is obtained " "from U = Q*X and V_T = P_T * Y_T. Constructing a " "testmatrix A is done the reverse way starting from orthogonal " "matrices and a list of singular values. " "Each of these matrices can be dumped to screen."); prm.declare_entry ("Resulting matrices to print", "Sigma", ::Patterns::MultipleSelection("A|Q|B|P_T|X|Sigma|Y_T|U|V_T"), "Possible values: A,Q,B,P_T,X,Sigma,Y_T,U,V_T " " Entries must be separated by a colon." " " "In the same spirit as \"Original matrices to print\". " "Print what the factorization has found."); prm.leave_subsection(); prm.enter_subsection("runtime parameters"); prm.declare_entry("Number of components", "4", ::Patterns::Integer(), "Number of components"); prm.declare_entry("Maximal number of iterations", "6", ::Patterns::Integer(), "Maximal number of iterations"); prm.leave_subsection(); } void step5::PCATestUIParams::get(::ParameterHandler &prm) { this->TestUIParamsBase::get(prm);
Die Masse der Parameter dient dazu, sich die Details der Ergebnisse anschauen zu koennen.
prm.enter_subsection("Visualization flags."); this->possible_matrices_to_print_orig = QString("A|Q|B|P_T|X|Sigma|Y_T|U|V_T").split("|"); this->possible_matrices_to_print_num = this->possible_matrices_to_print_orig; QString parse_results = QString::fromStdString(prm.get ("Original matrices to print")); this->selected_orig = parse_results.replace(QRegExp(" "), "").split(","); parse_results = QString::fromStdString(prm.get ("Resulting matrices to print")); this->selected_num = parse_results.replace(QRegExp(" "), "").split(","); prm.leave_subsection(); prm.enter_subsection("runtime parameters"); this->n_components = prm.get_integer("Number of components"); this->max_iter = prm.get_integer("Maximal number of iterations"); prm.leave_subsection(); } template<typename T> void step5::PCATestUIParams::parse_wave_forms(::FullMatrix<T> & A) { std::vector<std::vector<T> > parsed_data; QFile file("mea-data/sp.dat");
single-electrode-single-neuron.dat"); wave-form-1.txt");
if (!file.open(QIODevice::ReadOnly | QIODevice::Text)) { QMessageBox mea_read_failed_message; mea_read_failed_message.setText("Reading MEA data has failed. " "Please retry manually."); QString filename = QFileDialog::getOpenFileName(0, QObject::tr("Retry opening MEA Data File"), ".", QObject::tr("MEA time series (*.dat)"));
Check, whether opening the file worked. If not Something must have gone terribly wrong. Thus: Give up.
file.setFileName(filename);
if (!file.open(QIODevice::ReadOnly | QIODevice::Text))
AssertThrow(false,
::ExcMessage("Could not read any MEA data. Giving up."));
}
else
std::cout
<< "mea-data/single-electrode-single-neuron.dat successfully opened."
<< "mea-data/sp.dat sucessfully opened" << std::endl; unsigned int line_length = 32; //Gazaleh: 75; unsigned int n_rows = 0;
count rows first. Otherwise we run into troulbe with large data sets.
{
QTextStream line_counter(&file);
while (!line_counter.atEnd())
{
line_counter.readLine();
n_rows++;
}
}
std::cout
<< "n_rows : " << n_rows << ", n_cols : "
<< line_length << std::endl;
A.reinit(n_rows, line_length);
QTextStream in(&file);
reset to the beginning of the stream, i.e. the first line.
in.seek(0);
unsigned int row = 0; // row counter
while (!in.atEnd())
{
QString line = in.readLine();
QStringList linedata = line.split(" ");
std::cout << "Row " << row << line.toStdString() << std::endl;
for (unsigned int i = 0; i < line_length; ++i) A(row,i) = T(linedata[i].toDouble()); row++; } std::cout << "Parsing wave forms done" << std::endl; }
------------------------------
template <typename T, typename blas> step5::PCATest<T, blas>::PCATest(::ParameterHandler &prm) : n_successful_measurements(0) { params.get(prm); } template <typename T, typename blas> void step5::PCATest<T, blas>::run() {
setup_and_assemble(1024, 128+32*8); // this->params.max_n_cols);
for (int nr = params.min_n_cols; nr <= params.max_n_cols; ) { for (int nc = params.min_n_cols; nc <= nr; nc *= params.n_rows_growth_rate) { setup_and_assemble(nr, nc); factorize(); } nr = std::max(nr + 1, int(nr * params.n_rows_growth_rate)); } save_results(); } template <typename T, typename blas> void step5::PCATest<T, blas>::factorize() { #ifdef USE_SYNTH_DATA ::FullMatrix<T> A(B.n_rows(), B.n_cols() ), tmp(B.n_rows(), B.n_cols() ); B.mmult(tmp, this->P_t); this->Q.mmult(A, tmp); #endif
CudaSVDecomposition<T, blas> SVD; SVD.factorize(A);
check_results(A, SVD);
SVD.factorize_pca(A);
check_results(A, SVD);
const unsigned int n_repetitions = 2; //this->params.n_repetitions; #ifndef USE_SYNTH_DATA ::FullMatrix<T> A; this->params.parse_wave_forms(A); #endif step4::CudaQRDecomposition<T, blas> QR; CUDATimer qr_timer; T elapsed_time = QR.householder(A); qr_timer.stop(); qr_timer.print_elapsed("Overall time for QR-preprocessing"); FullMatrixAccessor R_tmp; R_tmp = QR.R(); std::cout << "R : from QR-Preprocessing\n"; ::FullMatrix<T> R_tmp_h(R_tmp.n_cols(), R_tmp.n_cols()); std::ofstream R_out("R_mea.dat"); for (unsigned int r = 0; r < R_tmp_h.n_cols(); ++r) { for(unsigned int c = 0; c < R_tmp_h.n_cols(); ++c) { R_tmp_h(r,c) = R_tmp(r,c);
if (r == c)
R_out << QR.R()(r,c) << " "; } R_out << std::endl; } std::cout << std::endl; std::vector<::Vector<double> > times(2, ::Vector<double>(n_repetitions)); std::cout << "Nipals test for " << A.n_rows() << " rows, " << A.n_cols() << " cols :\n" << "----------------------------------------------------------" << std::endl; for (unsigned int k = 0; k < n_repetitions; k++) {
CudaTruncatedSVDecomposition<T, blas> tSVD;
tSVD.factorize(A); tSVD.Lambda().print();
CudaNIPALS<T, blas> nipals(params.n_components, params.max_iter);
times[0](k) = nipals.get_pca(R_tmp_h);
std::cout << "SV PCA++:\n";
nipals.lambda.print(std::cout);
times[1](k) = nipals.get_pca_blas_version(R_tmp_h);
std::cout << "SV PCA--:\n";
nipals.lambda.print(std::cout);
}
std::vector<double> mean(2), std_dev(2), rel_error(2);
::Vector<double> tmp_data(n_repetitions);
Da Laufzeiten immer positiv sind, entspricht die L1-norm im Wesentlichen dem Mittelwert.
for (unsigned int e = 0; e < 2; e++) { mean[e] = times[e].l1_norm()/n_repetitions; tmp_data = mean[e]; tmp_data -= times[e];
Instead of te plain standard deviation we rathe rmeasure the uncertainty of the average run-time which gives a tighter error bound.
std_dev[e] = tmp_data.l2_norm()/std::sqrt(n_repetitions);
if (n_repetitions > 1)
rel_error[e] = std_dev[e]/(mean[e]*std::sqrt(n_repetitions-1));
}
std::cout << "Statistics : \n"
<< "------------------------------------------------------------\n"
<< "PCA++ mean : " << mean[0] << ", rel error : " << rel_error[0] << "\n"
<< "PCA-blas mean : " << mean[1] << ", rel error : " << rel_error[1] << "\n"
<< "PCA++/PCA-blas : " << mean[0] / mean[1] << "\n"
<< "-----------------------------------------------------------\n"
<< std::endl;
If one of the relative errors is larger than 20% we skip the measurement. In ORIGIN: remove bad data points.
if (rel_error[0] <= .2 && rel_error[1] <= .2 )
{
Becuse of a design flaw in the ConvergenceTable class we have tokeep track of the number rows in the table by ourselves. If the table does not contain at least a single row the program crashes due to a seg fault when writing the table to disk.
this->n_successful_measurements++;
Speichere die mittleren Laufzeiten in der Ergebnis-Tabelle fuer spaeteres plotten.
this->results_table.add_value("n_rows ", A.n_rows() ); this->results_table.add_value("n_cols ", A.n_cols() ); this->results_table.add_value ("Laufzeit PCA++ / sec", mean[0] ); this->results_table.set_precision ("Laufzeit PCA++ / sec", 16); this->results_table.add_value ("std_dev PCA++ / sec", std_dev[0] ); this->results_table.set_precision ("std_dev PCA++ / sec", 16); this->results_table.add_value ("uncertainty PCA++ / sec", std_dev[0]/std::sqrt(n_repetitions-1) ); this->results_table.set_precision ("uncertainty PCA++ / sec", 16); this->results_table.add_value ("Laufzeit blas PCA / sec", mean[1] ); this->results_table.set_precision ("Laufzeit blas PCA / sec", 16); this->results_table.add_value ("std_dev blas PCA / sec", std_dev[1] ); this->results_table.set_precision ("std_dev blas PCA / sec", 16); this->results_table.add_value ("uncertainty PCA / sec", std_dev[1]/std::sqrt(n_repetitions-1) ); this->results_table.set_precision ("uncertainty PCA / sec", 16); } } template <typename T, typename blas> void step5::PCATest<T, blas>::setup_and_assemble(unsigned int nr, unsigned int nc) { Assert(nr >= nc, ::ExcMessage("n_cols must not exceed n_rows.")); MatrixCreator::extended_normalized_hadamard(nr, this->H); this->Q.reinit(nr, nr); for (unsigned int r = 0; r < nr; ++r) for (unsigned int c = 0; c < nr; ++c) Q(r,c) = H(r,c); MatrixCreator::extended_normalized_hadamard(nc, this->H); this->P_t.reinit(nc, nc); for (unsigned int r = 0; r < nc; ++r) for (unsigned int c = 0; c < nc; ++c) P_t(r,c) = H(r,c);
Die zu rekonstruierende obere Bidiagonalmatrix wird zeilenweise fortlaufend mit 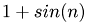 ,
, 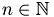 gefuellt, wodurch die Matrixeintraege saemtlich in dem Intervall
gefuellt, wodurch die Matrixeintraege saemtlich in dem Intervall 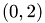 liegen.
liegen.
this->B.reinit(nr, nc);
int tmp = 1;
for (unsigned int r = 0; r < nc; ++r)
{
B(r, r) = 1+std::sin(tmp++);
if (r < nc-1)
B(r, r+1) = -1; // + std::sin(tmp++);
}
bool print_extra_blank_line = false;
Bei Bedarf werden die Originalmatrizen ausgegeben.
if (this->params.selected_orig.contains("Q") ) { std::cout << "Initial Q : \n"; std::cout << "-------------------------------------------------" << std::endl; Q.print(std::cout); print_extra_blank_line = true; } if (this->params.selected_orig.contains("B") ) { std::cout << "\nInitial B : \n"; std::cout << "-------------------------------------------------" << std::endl; B.print(std::cout); print_extra_blank_line = true; } if (this->params.selected_orig.contains("P_T") ) { std::cout << "Initial P_t : \n"; std::cout << "-------------------------------------------------" << std::endl; P_t.print(std::cout); print_extra_blank_line = true; } if (print_extra_blank_line) std::cout << std::endl; } template <typename T, typename blas> void step5::PCATest<T, blas>::check_results(const ::FullMatrix<T> & / *A* /, const CudaNIPALS<T, blas> & / *SVDf* /) { } template <typename T, typename blas> void step5::PCATest<T, blas>::save_results() { if (this->n_successful_measurements > 0) this->params.template save_results<T, blas>(std::string("NIPALSTest"), results_table); else std::cout << "------------------------------------------------------------\n" << " No data to write to disk. \n" << " Possible reason: Wall times fluctuated too much.\n" << " Please rerun the test.\n" << "------------------------------------------------------------\n" << std::endl; }
------------------------------
namespace step5 {
Function: run_pca_tests
Hier werden die Benutzereingaben verarbeitet und an die Tests uebergeben.
void run_pca_tests(int / *argc* /, char *argv[]) {
Declare and read parameters from a file. Basically, the parameter file must have the same name as the binary. The extension has to be ".prm". What has been read will be dumped into a log file.
::ParameterHandler prm_handler;
step4::SimUIParams::declare(prm_handler);
PCATestUIParams::declare(prm_handler);
std::string prm_filename(argv[0]);
prm_filename += "-PCA.prm";
prm_handler.read_input (prm_filename);
prm_filename += ".log";
std::ofstream log_out_text(prm_filename.c_str());
prm_handler.print_parameters (log_out_text,
::ParameterHandler::Text);
step4::SimUIParams params;
params.get(prm_handler);
for(int DevNo = 0; DevNo < global_data.n_CUDA_devices; DevNo++)
{
cudaSetDevice(DevNo);
global_data.current_device_id = DevNo;
global_data.cublanc_gpu_info();
if (params.run_cublas_float) {
std::cout << "Householder-QR<float> mit cublas :\n"
<< "------------------------------------------------" << std::endl;
Test fuer cublas
PCATest<float, cublas> cublas_pca_test_float(prm_handler);
cublas_pca_test_float.run();
std::cout << "\nHouseholder-QR<float> mit cublas DONE \n"
<< "------------------------------------------------\n" << std::endl;
}
Falls doppelte Genaugikeit zur Verfuegung steht, werden die Tests dafuer ausgefuehrt.
#ifdef CUDA_DOUBLE_CAPABLE if (params.run_cublas_double) { std::cout << "\nHouseholder-QR<double> mit cublas :\n" << "------------------------------------------------" << std::endl; PCATest<double, cublas> cublas_pca_test_double(prm_handler); cublas_pca_test_double.run(); std::cout << "\nHouseholder-QR<double> mit cublas DONE \n" << "------------------------------------------------\n" << std::endl; } #endif } if (params.run_cpu_blas_float) { std::cout << "\nHouseholder-QR<float> mit ATLAS/CBLAS :\n" << "------------------------------------------------" << std::endl;
Test fuer blas
PCATest<float, blas> blas_qr_test_float(prm_handler);
blas_qr_test_float.run();
std::cout << "\nHouseholder-QR<float> mit ATLAS/CBLAS DONE \n"
<< "------------------------------------------------\n" << std::endl;
}
if (params.run_cpu_blas_double) {
std::cout << "\nHouseholder-QR<double> mit ATLAS/CBLAS :\n"
<< "------------------------------------------------" << std::endl;
PCATest<double, blas> blas_qr_test_double(prm_handler);
blas_qr_test_double.run();
std::cout << "\nHouseholder-QR<double> mit ATLAS/CBLAS DONE \n"
<< "------------------------------------------------\n" << std::endl;
}
}
} // namespace step5 END
Function: main
Process user input and fire up the tests.
int main(int argc, char *argv[]) { cudaGetDeviceCount(&global_data.n_CUDA_devices); std::cout << "N available CUDA devices : " << global_data.n_CUDA_devices << std::endl; QApplication app(argc, argv); step5::run_pca_tests(argc, argv); std::cout << std::endl; return app.exec(); }
Results
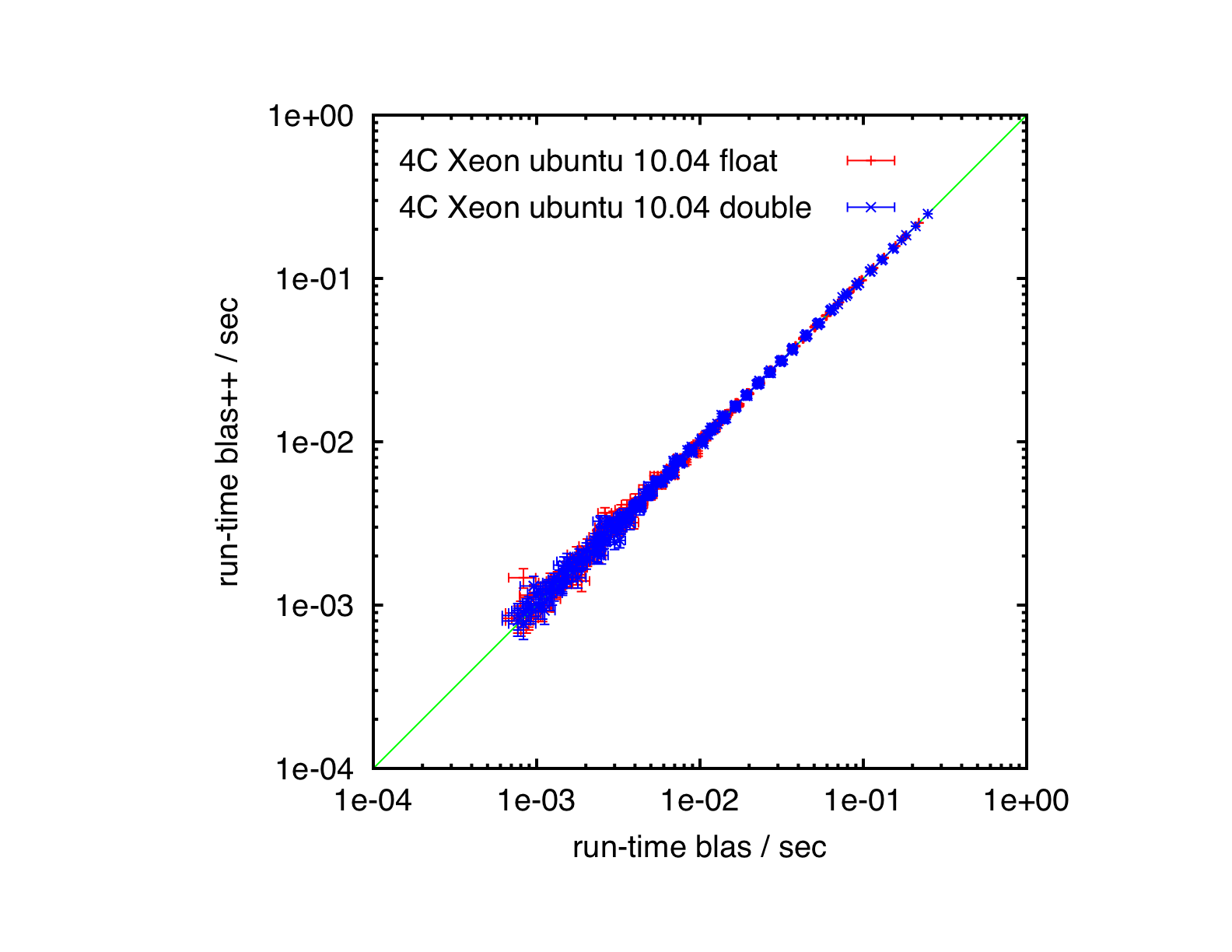
Quad-core Xeon, Ubuntu 10.04, ATLAS
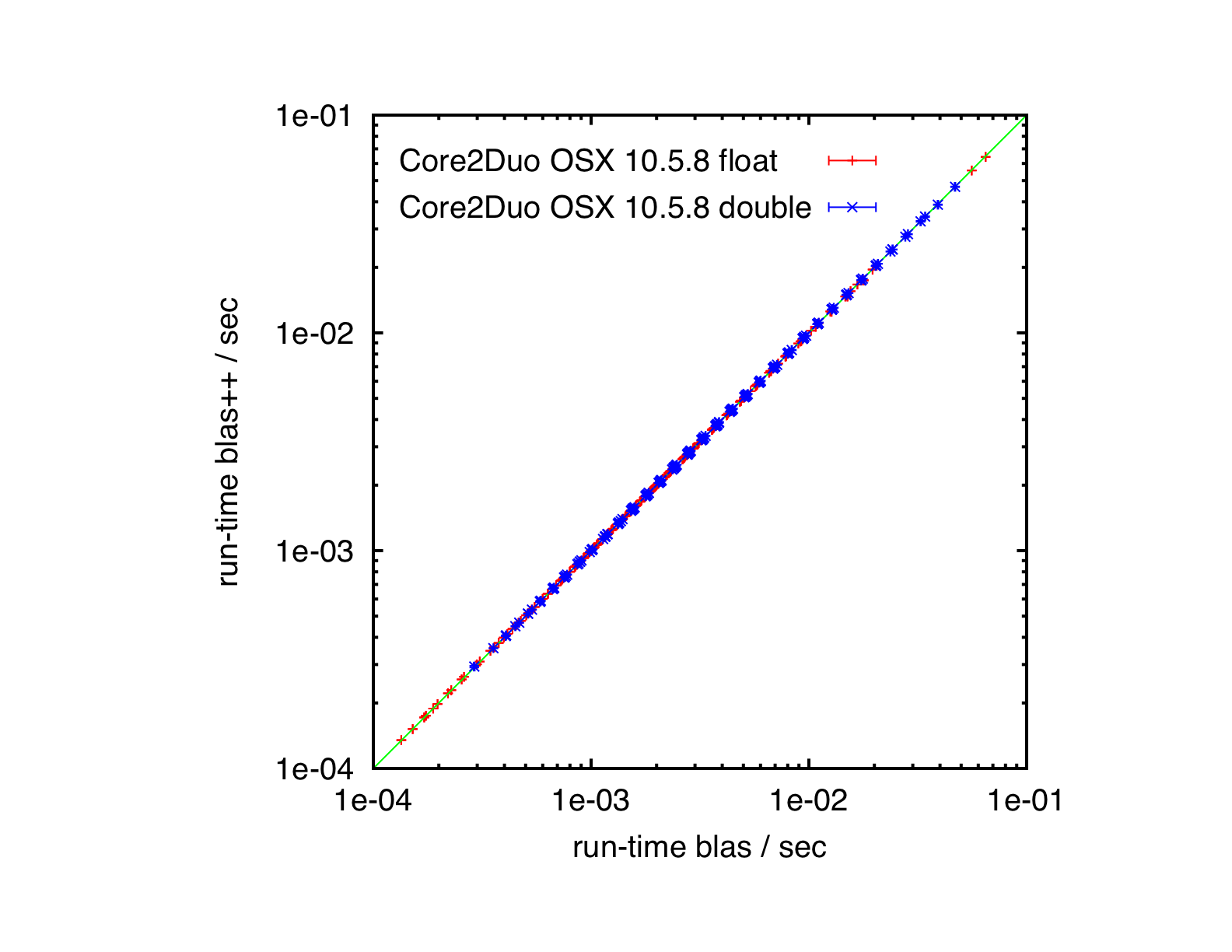
Core2Duo, Mac OS X 10.5.8, ATLAS
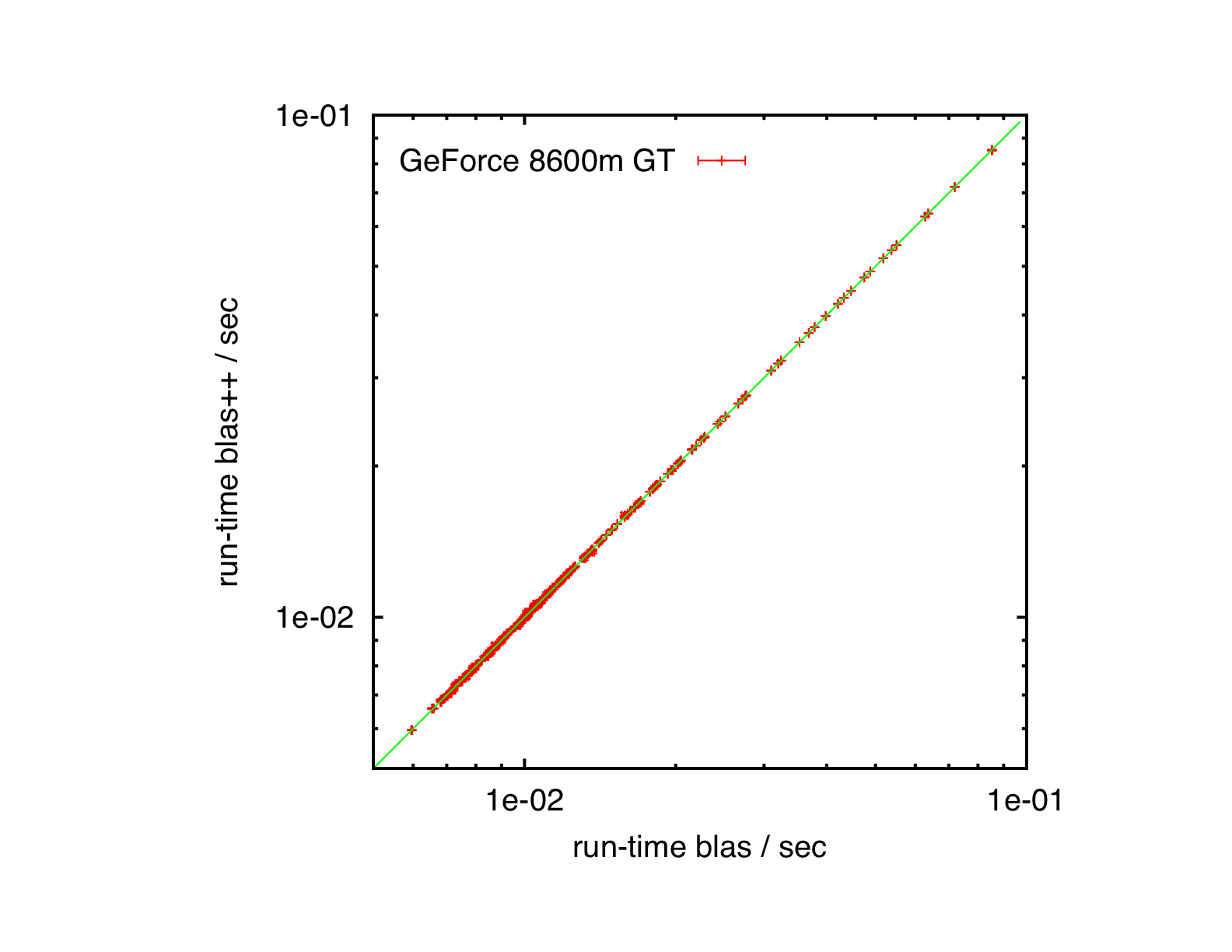
GeForce 8600m GT, Mac OS X 10.5.8, CUBLAS
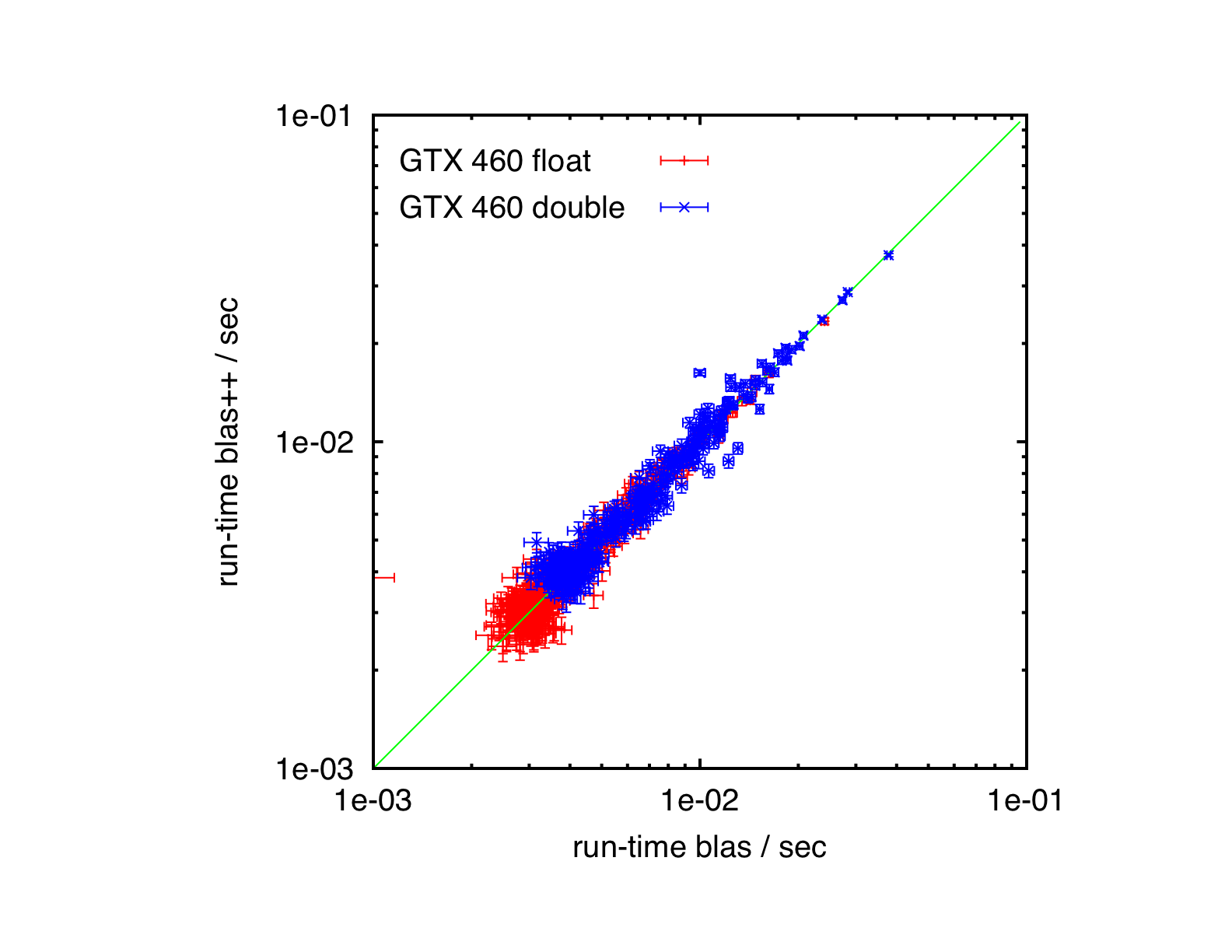
GeForce GTX 460, Ubuntu 10.04, CUBLAS
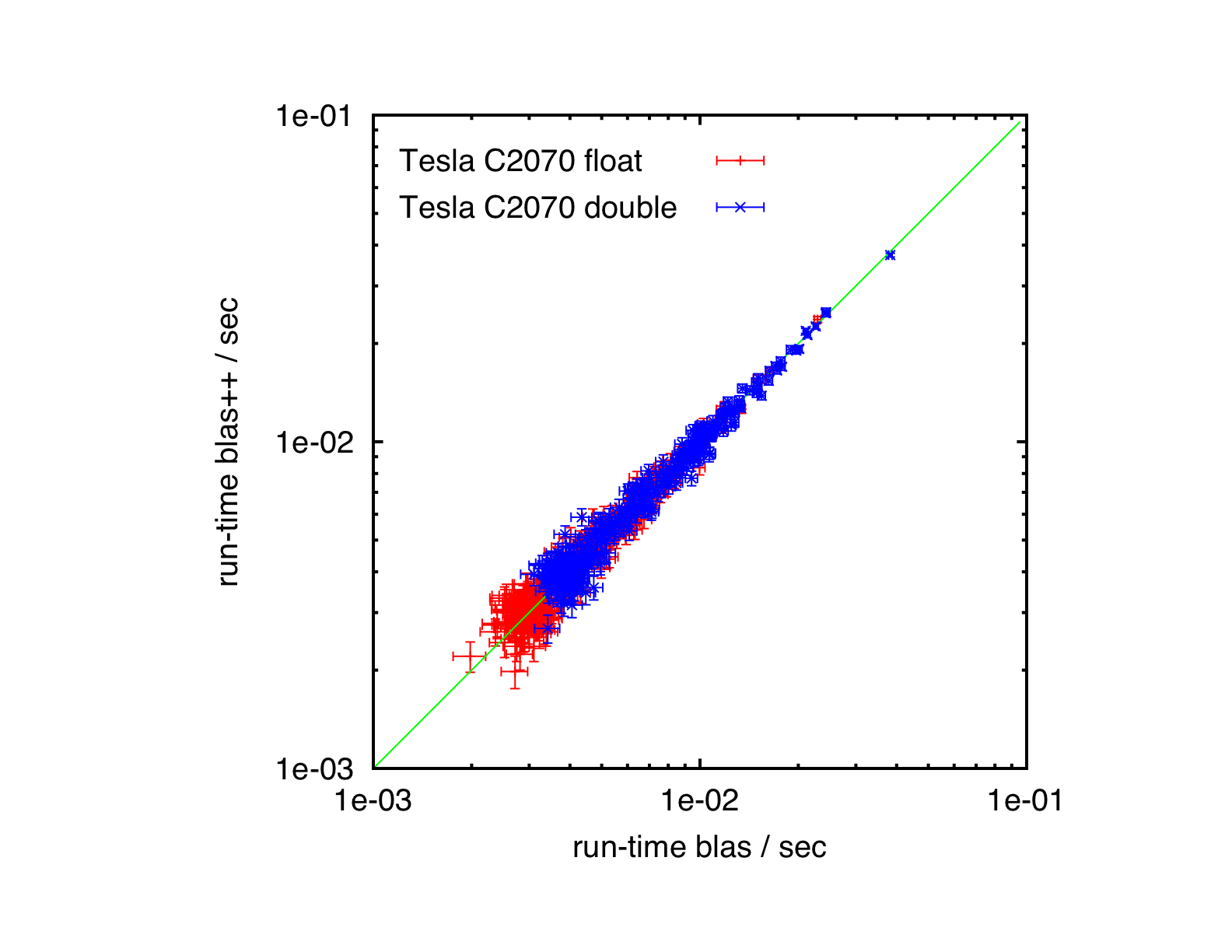
Tesla C2070, Ubuntu 10.04, CUBLAS
The plain program
(If you are looking at a locally installed CUDA HPC Praktikum version, then the program can be found at .. /.. /testsite / /step-5 /step-cu.cc . Otherwise, this is only the path on some remote server.)
#ifndef CUDA_KERNEL_STEP_5_CU_H #define CUDA_KERNEL_STEP_5_CU_H
void step_5_dummy(); #endif // CUDA_KERNEL_STEP_5_CU_H #include <cutil_inline.h> #include <cuda_kernel_wrapper_step-5.cu.h>
__device__ int lex_index_2D(int r, int c, int row_length) { return c + r*row_length; }
__global__ void single_thread_cholesky(float *A, const int gpu_tile_size) { typedef float T; for (unsigned int r = 0; r < gpu_tile_size; ++r) { T sum = 0.; unsigned int idx; unsigned int idx_c; for (unsigned int u = 0; u < r; ++u) { idx = lex_index_2D(r, u, gpu_tile_size); sum += A[idx] * A[idx]; } idx = lex_index_2D(r, r, gpu_tile_size); A[idx] = sqrt(A[idx] - sum); for (unsigned int c = r+1; c < gpu_tile_size; ++c) { sum = 0.; for (unsigned int u = 0; u < r; ++u) { idx_c = lex_index_2D(c, u, gpu_tile_size); idx = lex_index_2D(r, u, gpu_tile_size); sum += A[idx_c]*A[idx]; } idx_c = lex_index_2D(c, r, gpu_tile_size); idx = lex_index_2D(r, c, gpu_tile_size); A[idx_c] = A[idx] - sum; idx = lex_index_2D(r, r, gpu_tile_size); A[idx_c] /= A[idx]; } } }
__global__ void __step_5_dummy() { int i = 2; int j = 3; int k = i+j; i += k; }
void step_5_dummy() { dim3 grid(5,5); dim3 blocks(16,16); __step_5_dummy<<<grid, blocks>>>(); } #ifndef CUDADriver_STEP_5_H #define CUDADriver_STEP_5_H #include <lac/blas++.h> #include <lac/cublas_algorithms.h> #include <lac/full_matrix.h> namespace step5 {
template <typename T, typename blas> class CudaNIPALS { public: typedef typename blas_pp<T, blas>::blas_wrapper_type BW; typedef typename blas_pp<T, blas>::FullMatrixAccessor FullMatrixAccessor; typedef typename blas_pp<T, blas>::Matrix Matrix; typedef typename blas_pp<T, blas>::SubMatrix SubMatrix; typedef typename blas_pp<T, blas>::MatrixSubCol MatrixSubCol; typedef typename blas_pp<T, blas>::MatrixSubRow MatrixSubRow; typedef typename blas_pp<T, blas>::Vector Vector; CudaNIPALS(unsigned int p_n_components,unsigned int p_max_iter) : num_zero(1e-8),max_iter(p_max_iter), n_components(p_n_components) {} ~CudaNIPALS (); double get_pca (const ::FullMatrix<T>& X); double get_pca_blas_version (const ::FullMatrix<T>& X); const Matrix & scores() const { return T_d; } const Matrix & loads() const { return P_d; } const Matrix & residual() const { return R_d; } ::Vector<T> lambda; private: Matrix T_d, P_d, R_d; unsigned int n_components; //X.n_cols(); const double num_zero; // = 1e-8; unsigned int max_iter; };
template <typename T, typename blas> class CudaTruncatedSVDecomposition { public: typedef typename blas_pp<T, blas>::blas_wrapper_type BW; typedef typename blas_pp<T, blas>::FullMatrixAccessor FullMatrixAccessor; typedef typename blas_pp<T, blas>::Array Array; typedef typename blas_pp<T, blas>::Matrix Matrix; typedef typename blas_pp<T, blas>::SubMatrix SubMatrix; typedef typename blas_pp<T, blas>::MatrixSubCol MatrixSubCol; typedef typename blas_pp<T, blas>::MatrixSubRow MatrixSubRow; typedef typename blas_pp<T, blas>::Vector Vector; typedef typename bw_types::algorithms<T> algorithms; CudaTruncatedSVDecomposition() {} ~CudaTruncatedSVDecomposition() {} void factorize(const ::FullMatrix<T>& A); const Matrix & V() const { return U_d; } const Matrix & U() const { return V_d; } private: Matrix R_d, U_d, V_d; ::Vector<T> __Lambda; }; } // namespace step5 END #endif // CUDADriver_STEP_5_H #ifndef CUDA_DRIVER_STEP_5_HH #define CUDA_DRIVER_STEP_5_HH #include "cuda_driver_step-5.h" #include "cuda_kernel_wrapper_step-5.cu.h" #include <cutil_inline.h> #include <base/CUDATimer.h> #include <base/timer.h>
template <typename T, typename blas> step5::CudaNIPALS<T, blas>::~CudaNIPALS() {} static const bool measure_all = true; static const bool measure_RT_times_tk = false; static const bool measure_R_times_pk = false;
template <typename T, typename blas> double step5::CudaNIPALS<T, blas>::CudaNIPALS::get_pca(const ::FullMatrix<T>& X) { ::FullMatrixAccessor<T> X_t_ugly(X, true / *transpose copy* /); this->R_d = X_t_ugly; this->T_d.reinit(X.n_rows(), n_components ); this->P_d.reinit(X.n_cols(), n_components ); lambda.reinit(n_components); transpose<Matrix> R_T(this->R_d); MatrixSubCol hat_t_k (this->T_d, 0, 0); MatrixSubCol hat_p_k(this->P_d, 0, 0); MatrixSubCol hat_r_k(this->R_d, 0, 0); Vector p_k(hat_p_k.size() ), t_k(hat_t_k.size() ); double net_time = 0.; CUDATimer pca_timer; for (unsigned int k = 0; k < n_components; k++) { T lambda_old = 0.; lambda(k) = 0.; bool converged = false; hat_t_k.reset(0, k); hat_p_k.reset(0, k); hat_r_k.reset(0, k); t_k = hat_r_k; for (unsigned int j = 0; j < max_iter //&& !converged ; j++) { if (measure_RT_times_tk && !measure_all) { pca_timer.reset(); p_k = R_T * t_k; pca_timer.stop(); net_time += pca_timer(); } else p_k = R_T * t_k; p_k /= p_k.l2_norm(); if (measure_R_times_pk && !measure_all) { pca_timer.reset(); t_k = R_d * p_k; pca_timer.stop(); net_time += pca_timer(); } else t_k = R_d * p_k; lambda(k) = t_k.l2_norm(); converged = (std::fabs(lambda(k) - lambda_old) <= num_zero); lambda_old = lambda(k); } R_d.add_scaled_outer_product(-1., t_k, p_k); hat_p_k = p_k; hat_t_k = t_k; } if (measure_all && !(measure_RT_times_tk || measure_R_times_pk)) { pca_timer.stop(); net_time += pca_timer(); } return net_time; }
template <typename T, typename blas> double step5::CudaNIPALS<T, blas>::CudaNIPALS::get_pca_blas_version(const ::FullMatrix<T>& X) { ::FullMatrixAccessor<T> X_t_ugly(X, true / *Erstelle Kopie der Transponierten* /); this->R_d = X_t_ugly; this->P_d.reinit(n_components, X.n_cols() ); this->T_d.reinit(X.n_rows(), n_components ); this->lambda.reinit(n_components); double net_time = 0.; CUDATimer pca_timer; int M = this->R_d.n_rows(); int N = this->R_d.n_cols(); int K = n_components; T * dR = this->R_d.array().val(); T * dT = this->T_d.array().val(); T * dP = this->P_d.array().val(); for(int k=0; k < K; k++) { BW::copy(M, &dR[k*M], 1, &dT[k*M], 1); T lambda_old = 0.; for(unsigned int j=0; j < max_iter; j++) { if (measure_RT_times_tk && !measure_all) { pca_timer.reset(); BW::gemv('t', M, N, 1.0, dR, M, &dT[k*M], 1, 0.0, &dP[k*N], 1); pca_timer.stop(); net_time += pca_timer(); } else BW::gemv('t', M, N, 1.0, dR, M, &dT[k*M], 1, 0.0, &dP[k*N], 1); BW::scal(N, 1.0/BW::nrm2(N, &dP[k*N], 1), &dP[k*N], 1); if (measure_R_times_pk && !measure_all) { pca_timer.reset(); BW::gemv('n', M, N, 1.0, dR, M, &dP[k*N], 1, 0.0, &dT[k*M], 1); pca_timer.stop(); net_time += pca_timer(); } else BW::gemv('n', M, N, 1.0, dR, M, &dP[k*N], 1, 0.0, &dT[k*M], 1); lambda(k) = BW::nrm2(M, &dT[k*M], 1); if(fabs(lambda_old - lambda(k)) < num_zero) break; lambda_old = lambda(k); } BW::ger(M, N, -1.0, &dT[k*M], 1, &dP[k*N], 1, dR, M); } if (measure_all && !(measure_RT_times_tk || measure_R_times_pk)) { pca_timer.stop(); net_time += pca_timer(); } return net_time; } template <typename T, typename blas> void step5::CudaTruncatedSVDecomposition<T, blas>::factorize(const ::FullMatrix<T>& A) { ::FullMatrixAccessor<T> A_t_ugly(A, true / *Erstelle Kopie der Transponierten* /); this->R_d = A_t_ugly; const unsigned int n_components = A.n_cols(); static const unsigned int max_iter = 1000; static const T num_zero = 1e-5; this->V_d.reinit(A.n_rows(), n_components); this->U_d.reinit(A.n_cols(), n_components); this->__Lambda.reinit (n_components); transpose<Matrix> R_T(R_d); SubMatrix V_k(V_d, 0, 0); transpose<SubMatrix> V_k_T(V_k); SubMatrix U_k(U_d, 0, 0); transpose<SubMatrix> U_k_T(U_k); Vector v_k (A.n_rows()); Vector b (A.n_rows()); Vector u_k (A.n_cols()); Vector a (A.n_cols()); int r_begin = 0; int r_end_U = U_d.n_rows(); int r_end_V = V_d.n_rows(); for (unsigned int k = 0; k < n_components; k++) { T lambda = 0., lambda_old = 0.; MatrixSubCol v_k_view(R_d, 0, k), u_k_view(U_d, 0, k), r_k(R_d, 0, k); v_k = r_k; bool converged = false; int new_c_begin = 0; int new_c_end = k; if (k>0) { U_k.reset(r_begin, r_end_U, new_c_begin, new_c_end); V_k.reset(r_begin, r_end_V, new_c_begin, new_c_end); } for (unsigned int j = 0; j < max_iter && !converged; j++) { u_k = R_T * v_k; #define USE_REORTHO #ifdef USE_REORTHO if (k > 0) { a = U_k_T * u_k; u_k -= U_k * a; } #endif u_k /= u_k.l2_norm(); v_k = R_d * u_k; #ifdef USE_REORTHO if (k > 0) { b = V_k_T * v_k; v_k -= V_k * b; } #endif lambda = v_k.l2_norm(); v_k /= lambda; converged = std::fabs(lambda -lambda_old) < num_zero; lambda_old = lambda; } R_d.add_scaled_outer_product(-lambda, v_k, u_k); v_k_view = v_k; u_k_view = u_k; this->__Lambda(k) = lambda; } std::cout << "SV from tSVD : \n"; this->__Lambda.print(std::cout, 8, false); } #endif // CUDA_DRIVER_STEP_5_HH #include <iostream> #include <vector> #include <QApplication> #include <QString> #include <QMessageBox> #include <QFileDialog> #include "cuda_driver_step-5.h" #include "cuda_driver_step-5.hh" #include "../step-4/step-4.hh" namespace step5 {
struct PCATestUIParams : public step4::TestUIParamsBase { PCATestUIParams() : TestUIParamsBase() {} static void declare(::ParameterHandler & prm); void get(::ParameterHandler & prm); QStringList possible_matrices_to_print_orig; QStringList selected_orig; QStringList possible_matrices_to_print_num; QStringList selected_num; unsigned int n_components, max_iter; template<typename T> void parse_wave_forms(::FullMatrix<T> & A); private: PCATestUIParams (const PCATestUIParams & / *other* /) : TestUIParamsBase() {} PCATestUIParams & operator = (const PCATestUIParams & / *other* /) { return *this; } }; template <typename T, typename blas> class PCATest { public: typedef typename blas_pp<T, blas>::FullMatrixAccessor FullMatrixAccessor; typedef typename blas_pp<T, blas>::Matrix Matrix; typedef transpose<Matrix> tr; typedef typename blas_pp<T, blas>::SubMatrix SubMatrix; typedef typename blas_pp<T, blas>::MatrixSubCol MatrixSubCol; typedef typename blas_pp<T, blas>::Vector Vector; PCATest(::ParameterHandler & prm); void run(); private: void setup_and_assemble(unsigned int nr, unsigned int nc); void factorize(); void check_results(const ::FullMatrix<T> & A, const CudaNIPALS<T, blas>& PCAf); void save_results(); ::FullMatrix<T> Q, B, P_t, H; PCATestUIParams params; ::ConvergenceTable results_table; unsigned int n_successful_measurements; }; } void step5::PCATestUIParams::declare(::ParameterHandler &prm) { TestUIParamsBase::declare(prm); prm.enter_subsection("Visualization flags."); prm.declare_entry ("Original matrices to print", "A", ::Patterns::MultipleSelection("A|Q|B|P_T|X|Sigma|Y_T|U|V_T"), "Possible values: A,Q,B,P_T,X,Sigma,Y_T,U,V_T " " Entries must be separated by a colon." " " "The direct SVD is a two-stage process. " "In the first factorization gives A = Q * B * P_T, " "where B is a upper bidiagonal matrix, and Q and P_T " "are orthogonal matrices. In the second factorization is " "B = X * Sigma * Y_T with S diagonal, containing the " "singular values and X and Y_T are orthogonal matrices again. " "The final result A = U * Sigma * V_T of the SVD is obtained " "from U = Q*X and V_T = P_T * Y_T. Constructing a " "testmatrix A is done the reverse way starting from orthogonal " "matrices and a list of singular values. " "Each of these matrices can be dumped to screen."); prm.declare_entry ("Resulting matrices to print", "Sigma", ::Patterns::MultipleSelection("A|Q|B|P_T|X|Sigma|Y_T|U|V_T"), "Possible values: A,Q,B,P_T,X,Sigma,Y_T,U,V_T " " Entries must be separated by a colon." " " "In the same spirit as \"Original matrices to print\". " "Print what the factorization has found."); prm.leave_subsection(); prm.enter_subsection("runtime parameters"); prm.declare_entry("Number of components", "4", ::Patterns::Integer(), "Number of components"); prm.declare_entry("Maximal number of iterations", "6", ::Patterns::Integer(), "Maximal number of iterations"); prm.leave_subsection(); } void step5::PCATestUIParams::get(::ParameterHandler &prm) { this->TestUIParamsBase::get(prm); prm.enter_subsection("Visualization flags."); this->possible_matrices_to_print_orig = QString("A|Q|B|P_T|X|Sigma|Y_T|U|V_T").split("|"); this->possible_matrices_to_print_num = this->possible_matrices_to_print_orig; QString parse_results = QString::fromStdString(prm.get ("Original matrices to print")); this->selected_orig = parse_results.replace(QRegExp(" "), "").split(","); parse_results = QString::fromStdString(prm.get ("Resulting matrices to print")); this->selected_num = parse_results.replace(QRegExp(" "), "").split(","); prm.leave_subsection(); prm.enter_subsection("runtime parameters"); this->n_components = prm.get_integer("Number of components"); this->max_iter = prm.get_integer("Maximal number of iterations"); prm.leave_subsection(); } template<typename T> void step5::PCATestUIParams::parse_wave_forms(::FullMatrix<T> & A) { std::vector<std::vector<T> > parsed_data; QFile file("mea-data/sp.dat"); if (!file.open(QIODevice::ReadOnly | QIODevice::Text)) { QMessageBox mea_read_failed_message; mea_read_failed_message.setText("Reading MEA data has failed. " "Please retry manually."); QString filename = QFileDialog::getOpenFileName(0, QObject::tr("Retry opening MEA Data File"), ".", QObject::tr("MEA time series (*.dat)")); file.setFileName(filename); if (!file.open(QIODevice::ReadOnly | QIODevice::Text)) AssertThrow(false, ::ExcMessage("Could not read any MEA data. Giving up.")); } else std::cout << "mea-data/sp.dat sucessfully opened" << std::endl; unsigned int line_length = 32; //Gazaleh: 75; unsigned int n_rows = 0; { QTextStream line_counter(&file); while (!line_counter.atEnd()) { line_counter.readLine(); n_rows++; } } std::cout << "n_rows : " << n_rows << ", n_cols : " << line_length << std::endl; A.reinit(n_rows, line_length); QTextStream in(&file); in.seek(0); unsigned int row = 0; // row counter while (!in.atEnd()) { QString line = in.readLine(); QStringList linedata = line.split(" "); for (unsigned int i = 0; i < line_length; ++i) A(row,i) = T(linedata[i].toDouble()); row++; } std::cout << "Parsing wave forms done" << std::endl; } template <typename T, typename blas> step5::PCATest<T, blas>::PCATest(::ParameterHandler &prm) : n_successful_measurements(0) { params.get(prm); } template <typename T, typename blas> void step5::PCATest<T, blas>::run() { for (int nr = params.min_n_cols; nr <= params.max_n_cols; ) { for (int nc = params.min_n_cols; nc <= nr; nc *= params.n_rows_growth_rate) { setup_and_assemble(nr, nc); factorize(); } nr = std::max(nr + 1, int(nr * params.n_rows_growth_rate)); } save_results(); } template <typename T, typename blas> void step5::PCATest<T, blas>::factorize() { #ifdef USE_SYNTH_DATA ::FullMatrix<T> A(B.n_rows(), B.n_cols() ), tmp(B.n_rows(), B.n_cols() ); B.mmult(tmp, this->P_t); this->Q.mmult(A, tmp); #endif const unsigned int n_repetitions = 2; //this->params.n_repetitions; #ifndef USE_SYNTH_DATA ::FullMatrix<T> A; this->params.parse_wave_forms(A); #endif step4::CudaQRDecomposition<T, blas> QR; CUDATimer qr_timer; T elapsed_time = QR.householder(A); qr_timer.stop(); qr_timer.print_elapsed("Overall time for QR-preprocessing"); FullMatrixAccessor R_tmp; R_tmp = QR.R(); std::cout << "R : from QR-Preprocessing\n"; ::FullMatrix<T> R_tmp_h(R_tmp.n_cols(), R_tmp.n_cols()); std::ofstream R_out("R_mea.dat"); for (unsigned int r = 0; r < R_tmp_h.n_cols(); ++r) { for(unsigned int c = 0; c < R_tmp_h.n_cols(); ++c) { R_tmp_h(r,c) = R_tmp(r,c); R_out << QR.R()(r,c) << " "; } R_out << std::endl; } std::cout << std::endl; std::vector<::Vector<double> > times(2, ::Vector<double>(n_repetitions)); std::cout << "Nipals test for " << A.n_rows() << " rows, " << A.n_cols() << " cols :\n" << "----------------------------------------------------------" << std::endl; for (unsigned int k = 0; k < n_repetitions; k++) { CudaNIPALS<T, blas> nipals(params.n_components, params.max_iter); times[0](k) = nipals.get_pca(R_tmp_h); std::cout << "SV PCA++:\n"; nipals.lambda.print(std::cout); times[1](k) = nipals.get_pca_blas_version(R_tmp_h); std::cout << "SV PCA--:\n"; nipals.lambda.print(std::cout); } std::vector<double> mean(2), std_dev(2), rel_error(2); ::Vector<double> tmp_data(n_repetitions); for (unsigned int e = 0; e < 2; e++) { mean[e] = times[e].l1_norm()/n_repetitions; tmp_data = mean[e]; tmp_data -= times[e]; std_dev[e] = tmp_data.l2_norm()/std::sqrt(n_repetitions); if (n_repetitions > 1) rel_error[e] = std_dev[e]/(mean[e]*std::sqrt(n_repetitions-1)); } std::cout << "Statistics : \n" << "------------------------------------------------------------\n" << "PCA++ mean : " << mean[0] << ", rel error : " << rel_error[0] << "\n" << "PCA-blas mean : " << mean[1] << ", rel error : " << rel_error[1] << "\n" << "PCA++/PCA-blas : " << mean[0] / mean[1] << "\n" << "-----------------------------------------------------------\n" << std::endl; if (rel_error[0] <= .2 && rel_error[1] <= .2 ) { this->n_successful_measurements++; this->results_table.add_value("n_rows ", A.n_rows() ); this->results_table.add_value("n_cols ", A.n_cols() ); this->results_table.add_value ("Laufzeit PCA++ / sec", mean[0] ); this->results_table.set_precision ("Laufzeit PCA++ / sec", 16); this->results_table.add_value ("std_dev PCA++ / sec", std_dev[0] ); this->results_table.set_precision ("std_dev PCA++ / sec", 16); this->results_table.add_value ("uncertainty PCA++ / sec", std_dev[0]/std::sqrt(n_repetitions-1) ); this->results_table.set_precision ("uncertainty PCA++ / sec", 16); this->results_table.add_value ("Laufzeit blas PCA / sec", mean[1] ); this->results_table.set_precision ("Laufzeit blas PCA / sec", 16); this->results_table.add_value ("std_dev blas PCA / sec", std_dev[1] ); this->results_table.set_precision ("std_dev blas PCA / sec", 16); this->results_table.add_value ("uncertainty PCA / sec", std_dev[1]/std::sqrt(n_repetitions-1) ); this->results_table.set_precision ("uncertainty PCA / sec", 16); } } template <typename T, typename blas> void step5::PCATest<T, blas>::setup_and_assemble(unsigned int nr, unsigned int nc) { Assert(nr >= nc, ::ExcMessage("n_cols must not exceed n_rows.")); MatrixCreator::extended_normalized_hadamard(nr, this->H); this->Q.reinit(nr, nr); for (unsigned int r = 0; r < nr; ++r) for (unsigned int c = 0; c < nr; ++c) Q(r,c) = H(r,c); MatrixCreator::extended_normalized_hadamard(nc, this->H); this->P_t.reinit(nc, nc); for (unsigned int r = 0; r < nc; ++r) for (unsigned int c = 0; c < nc; ++c) P_t(r,c) = H(r,c); this->B.reinit(nr, nc); int tmp = 1; for (unsigned int r = 0; r < nc; ++r) { B(r, r) = 1+std::sin(tmp++); if (r < nc-1) B(r, r+1) = -1; // + std::sin(tmp++); } bool print_extra_blank_line = false; if (this->params.selected_orig.contains("Q") ) { std::cout << "Initial Q : \n"; std::cout << "-------------------------------------------------" << std::endl; Q.print(std::cout); print_extra_blank_line = true; } if (this->params.selected_orig.contains("B") ) { std::cout << "\nInitial B : \n"; std::cout << "-------------------------------------------------" << std::endl; B.print(std::cout); print_extra_blank_line = true; } if (this->params.selected_orig.contains("P_T") ) { std::cout << "Initial P_t : \n"; std::cout << "-------------------------------------------------" << std::endl; P_t.print(std::cout); print_extra_blank_line = true; } if (print_extra_blank_line) std::cout << std::endl; } template <typename T, typename blas> void step5::PCATest<T, blas>::check_results(const ::FullMatrix<T> & / *A* /, const CudaNIPALS<T, blas> & / *SVDf* /) { } template <typename T, typename blas> void step5::PCATest<T, blas>::save_results() { if (this->n_successful_measurements > 0) this->params.template save_results<T, blas>(std::string("NIPALSTest"), results_table); else std::cout << "------------------------------------------------------------\n" << " No data to write to disk. \n" << " Possible reason: Wall times fluctuated too much.\n" << " Please rerun the test.\n" << "------------------------------------------------------------\n" << std::endl; } namespace step5 {
void run_pca_tests(int / *argc* /, char *argv[]) { ::ParameterHandler prm_handler; step4::SimUIParams::declare(prm_handler); PCATestUIParams::declare(prm_handler); std::string prm_filename(argv[0]); prm_filename += "-PCA.prm"; prm_handler.read_input (prm_filename); prm_filename += ".log"; std::ofstream log_out_text(prm_filename.c_str()); prm_handler.print_parameters (log_out_text, ::ParameterHandler::Text); step4::SimUIParams params; params.get(prm_handler); for(int DevNo = 0; DevNo < global_data.n_CUDA_devices; DevNo++) { cudaSetDevice(DevNo); global_data.current_device_id = DevNo; global_data.cublanc_gpu_info(); if (params.run_cublas_float) { std::cout << "Householder-QR<float> mit cublas :\n" << "------------------------------------------------" << std::endl; PCATest<float, cublas> cublas_pca_test_float(prm_handler); cublas_pca_test_float.run(); std::cout << "\nHouseholder-QR<float> mit cublas DONE \n" << "------------------------------------------------\n" << std::endl; } #ifdef CUDA_DOUBLE_CAPABLE if (params.run_cublas_double) { std::cout << "\nHouseholder-QR<double> mit cublas :\n" << "------------------------------------------------" << std::endl; PCATest<double, cublas> cublas_pca_test_double(prm_handler); cublas_pca_test_double.run(); std::cout << "\nHouseholder-QR<double> mit cublas DONE \n" << "------------------------------------------------\n" << std::endl; } #endif } if (params.run_cpu_blas_float) { std::cout << "\nHouseholder-QR<float> mit ATLAS/CBLAS :\n" << "------------------------------------------------" << std::endl; PCATest<float, blas> blas_qr_test_float(prm_handler); blas_qr_test_float.run(); std::cout << "\nHouseholder-QR<float> mit ATLAS/CBLAS DONE \n" << "------------------------------------------------\n" << std::endl; } if (params.run_cpu_blas_double) { std::cout << "\nHouseholder-QR<double> mit ATLAS/CBLAS :\n" << "------------------------------------------------" << std::endl; PCATest<double, blas> blas_qr_test_double(prm_handler); blas_qr_test_double.run(); std::cout << "\nHouseholder-QR<double> mit ATLAS/CBLAS DONE \n" << "------------------------------------------------\n" << std::endl; } } } // namespace step5 END
int main(int argc, char *argv[]) { cudaGetDeviceCount(&global_data.n_CUDA_devices); std::cout << "N available CUDA devices : " << global_data.n_CUDA_devices << std::endl; QApplication app(argc, argv); step5::run_pca_tests(argc, argv); std::cout << std::endl; return app.exec(); }
 documentation generated on Mon Jul 18 12:05:11 2011 by
doxygen
1.5.7
documentation generated on Mon Jul 18 12:05:11 2011 by
doxygen
1.5.7