The step-6 tutorial program
| Table of contents | |
|---|---|
Introduction
Automated Analysis of Multi-Electrode Array Measurements of Neurons
This step deals with automated spike sorting and cluster analysis of multi-electrode array measurements of neurons. Before starting the simulation, we have to decide if we want an off-line or an on-line method, since the off-line method gets the data as an input file containing all the relevant data. The on-line method on the contrary gets its data directly from the measurement i.e. as a stream. Therefore we need two different constructors, one for each method.
The data that we get from the measurement are a collection of waveforms in an 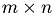 Matrix with each row being a waveform and the columns representing the time span of a single spike.
Matrix with each row being a waveform and the columns representing the time span of a single spike.
The wave forms are supposed to represent spikes generated from a few neurons in the vicinity of the electrode. It is assumed that each neuron produces a characteristic, typical waveform at the location of the electrode depending on parameters like electron-neuron-distances for instance.
The goal is to figure out (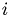 ) the shapes and number of these characteristic waveforms and (
) the shapes and number of these characteristic waveforms and (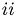 ) which measurement stems from which characteristic waveform.
) which measurement stems from which characteristic waveform.
To this end, each measurement is considered as a noisy representative of one of these characteristic waveforms so that, given a statistical model for the noise (i.e. a probability distribution for the deviations from the ideal waveforms) we are left with determining the parameters of this model.
This is a standard problem and can be solved by first computing a PCA of the data matrix (goal (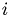 )) and then performing a cluster analysis, resolving goal (
)) and then performing a cluster analysis, resolving goal (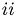 ).
).
The PCA works as follows: On the data matrix we use the QR decomposition as preprocessing step to reduce the amount of data. Here we have to denote, that using the QR decomposition as a preprocessor is not really efficient till 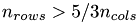 [8, p.253] which is the case in this project, since we have matrices with
[8, p.253] which is the case in this project, since we have matrices with 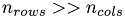 . We get the
. We get the 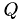 Matrix containing the data needed to reconstruct our initial matrix and the upper triangular matrix
Matrix containing the data needed to reconstruct our initial matrix and the upper triangular matrix 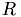 .
.
We can compute the principle component analysis of 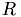 using e.g. the NIPALS algorithm to perform a truncated SVD, i.e. we decompose
using e.g. the NIPALS algorithm to perform a truncated SVD, i.e. we decompose 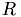 into orthogonal matrices
into orthogonal matrices 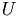 and
and 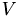 and the diagonal matrix
and the diagonal matrix 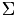 containing the singular values. The singular values describe the weight of a principal component.
containing the singular values. The singular values describe the weight of a principal component.
Using the matrices 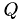 and
and 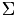 and the matrix
and the matrix 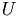 we construct a new Matrix
we construct a new Matrix 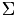 =
= 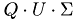 , on which we use a cluster analysis algorithm, for example K-means or Gaussian Mixture Models (GMM) to determine which neurons belong to which electrode.
, on which we use a cluster analysis algorithm, for example K-means or Gaussian Mixture Models (GMM) to determine which neurons belong to which electrode.
project flow
Both K-means and GMM are algorithms of the expectation-minimization type with an E-step being the expectation step, in which we get better point assignment and an M-step being the maximization step in which the model will be changed to be a better fit for the data points.
Purpose of this Step is using the K-means algorithm to determine the means of sample data generated within the program itself. The implementation of the K-means algorithm is based on the implementation in the Numerical Recipes [Third Edition] with the addition of OpenMP parallelization and also the implementation of the E-step of the algorithm as a cuda kernel.
Program design:
The cuda code is separated in host and device functions, the C code runs on the host (CPU) and the actual cuda code runs on the device (GPU). The host has to take care of the memory management of the device this includes allocations, deallocation and also copying data from and to the device. The actual device functions are called Kernels, for those we use kernel-wrapper functions to call them from inside the Host code. By using the kernel_wrapper, we can encapsulate the cuda code from the actual C++ code which allows us to use further programing technique such as template functions and also to be able to replace the cuda code whenever it is necessary, e.g. if we want to switch to a different parallelization technique.
Literature:
1. Spike sorting
2. Kmeans
3. K-means++: The Advantages of Careful Seeding
4. GMM
5. Robust, automatic spike sorting using mixtures of multivariate t-distributions. by Shy Shoham
6. QR Decomposition
7. NIPALS algorithm
8. Gene H. Golub and Charles F. Van Loan. Matrix Computations. Johns Hopkins University Press, 3rd edition, 1996.
The commented program
#include <iostream> #include <vector> #include <fstream> #include <QDir> #include <QString> #include <QVector> #include "Kmeans.h"
Function: main
This function handles the user inputs and pass them to the simulation
int main(int argc, char *argv[]) { cudaGetDeviceCount(&global_data.n_CUDA_devices); std::cout << "N available CUDA devices : " << global_data.n_CUDA_devices << std::endl;
run_tests(argc, argv);
} #ifndef CUDA_KERNEL_STEP_6_CU_H #define CUDA_KERNEL_STEP_6_CU_H
Declaration of Kernel-Wrapper-Functions
int e_step(int * cluster_assignment, int * counts, const float * data_points, float * cluster_centers, const int n_clusters, const int n_pca, const int n_data_points); #ifndef USE_SINGLE_PRECISION int e_step(int * cluster_assignment, int * counts, const double * data_points, double * cluster_centers, const int n_clusters, const int n_pca, const int n_data_points); #endif #endif // CUDA_KERNEL_STEP_6_CU_H
Header-File of the CUDA utility-Library.
#include <cutil_inline.h> #include <step-6/cuda_kernel_wrapper_step-6.cu.h>
Device Functions
Before the implementation of Kernels is discussed we need to implement some auxiliary functions. These "Device Functions" are only executable on the GPU itself and usually automatically inline although starting with compute capability 2.0 there is an additional keyword __force_inline__ to guarantee inlining.
Kernel: __e_step
The e-step of the K-means algorithm
template <typename T> __global__ void __e_step(int * cluster_assignment, const T * data_points, const T * cluster_centers, int * t_changes, const int n_clusters, const int n_pca, const int n_data_points, int * n_points_per_cluster) { int warp_id = threadIdx.x/ WARP_SIZE; int n_warps = (blockDim.x +WARP_SIZE -1)/ WARP_SIZE; int i = threadIdx.x + blockDim.x * blockIdx.x;
Starting with CUDA 3.2, shared memory can be allocated dynamically using malloc in a C-like fashion. In n_changes each warp tracks the number of changes in the assignment of data points to clusters.
__shared__ int * n_changes; if (threadIdx.x == 0) n_changes = (int*) malloc (WARP_SIZE*sizeof(int));
In counts we track the number of data_points assigned to each cluster in every thread block.
__shared__ int * counts; if (threadIdx.x == 0) counts = (int*) malloc (n_clusters*sizeof(int)); __syncthreads(); if(threadIdx.x < WARP_SIZE) n_changes[threadIdx.x] = 0; if(threadIdx.x < n_clusters) counts[threadIdx.x] = 0; __syncthreads(); if(i>=n_data_points) return; int c_min = 0, c_min_old = -1; T d_min = 0.; T tmp, distance_ij;
Measure the squared distance of this thread's data points to all cluster centers The first center is treated separately to properly initialize d_min
for(int d = 0; d < n_pca; d++) { tmp = data_points[n_pca*i+d]- cluster_centers[d]; d_min += tmp*tmp; c_min_old = cluster_assignment[i]; }
Measure the squared distance for the remaining clusters...
for(int c = 1; c < n_clusters; c++) { distance_ij = 0; for(int d = 0; d < n_pca; d++) { tmp = data_points[n_pca*i+d]- cluster_centers[n_pca*c+d]; distance_ij += tmp*tmp; }
... and check for minimal distance. Igf the new distance is smaller than the current minimum backup the distance id in c_min .
if(distance_ij < d_min)
{
d_min = distance_ij;
c_min = c;
}
}
cluster_assignment[i] = c_min;
increment the counts array using atomicAdd, because atomic operations are operations which are performed without interference from any other threads, and therefore prevent race conditions.
atomicAdd(counts+c_min, 1);
For each warp track the changes in the assigned cluster. The any function evaluates a predicate for all threads of the warp and returns non-zero if and only if the predicate evaluates to non-zero for any of them. Therefore we get a zero value if the cluster assignment doesn't change.
n_changes[warp_id] = abs(any(c_min - c_min_old));
__syncthreads();
if(threadIdx.x < n_clusters)
n_points_per_cluster[n_clusters*blockIdx.x+threadIdx.x] = counts[threadIdx.x];
if(threadIdx.x == 0)
{
t_changes[blockIdx.x] = 0;
for(int k = 0; k < n_warps; k++) t_changes[blockIdx.x] += n_changes[k];
__threadfence_block();
free(n_changes);
free(counts);
}
}
Kernel: __sum_counts_array
Sums up the counts vector.
Firstly the data in the counts vector are copied to shared memory to allow each thread to read from the array. The temp array in the shared memory is an array of arrays with the number of blocks in the X dimension and the number of clusters in the Y dimension. The idea behind this Kernel is to sum the data in each cluster from each block and if we get more data points than blocks, then we have to add the data from the array to the fields at the top. At the end we should have the complete array added in the Y first fields of the Array.
__global__ void __sum_counts_array(const int * n_points_per_cluster, const int n_data_points, int * counts, int * partialSum, unsigned int * counter) { const int ty = threadIdx.y; const int tx = threadIdx.x;
The bool variable isLastBlockDone is set to true if the Last Block is done calculating its sum. when this value is set to true we can calculate the last Sum of the count array.
__shared__ bool isLastBlockDone; if(tx == 0 && ty == 0) isLastBlockDone = false; __syncthreads(); #ifdef USE_ARRAY_OF_ARRAYS __shared__ int ** temp; if (tx == 0 && ty == 0) temp = (int**) malloc (blockDim.y*sizeof(int*)); __syncthreads(); if(tx == 0) temp[ty] = (int*) malloc (blockDim.x*sizeof(int)); __syncthreads(); temp[ty][threadIdx.x] = 0; __syncthreads();
The volatile keyword prevent the CUDA compiler from inline the operations needed to compute the value of a variable with volatile the complier is forced to really keep and reuse this variable in a register.
volatile int * myTemp = temp [ty] + tx; #else #ifdef USE_SHM_MALLOC __shared__ int * temp; if (tx == 0 && ty == 0) temp = (int*) malloc (blockDim.x*blockDim.y*sizeof(int)); #else __shared__ int temp[256 / *fixed size. thread blocks must not be larger! * / ]; #endif __syncthreads(); volatile int * myTemp = temp + blockDim.x*ty + tx; *myTemp = 0; __syncthreads(); #endif int global_index = ty + blockDim.y * tx + (blockDim.x*blockDim.y*blockIdx.x); if(global_index >= n_data_points) return; * myTemp = n_points_per_cluster[global_index]; __syncthreads(); for(int i = global_index + gridDim.x * blockDim.x*blockDim.y; i < n_data_points; i+= gridDim.x* blockDim.x*blockDim.y) { *myTemp += n_points_per_cluster[i]; __syncthreads(); } for(int k = blockDim.x/2; k > 0; k/=2) { if(tx < k) { *myTemp += *(myTemp + k); __syncthreads(); } } if(tx == 0) { partialSum[blockIdx.x * blockDim.y + ty] = *myTemp; __threadfence();
atomicInc checks if the Value at the address counter[ty] > = gridDim.x, if that is not the case, it increments the value at counter[ty] and stores the result back to global memory at the same address.
unsigned int value = atomicInc(&counter[ty], gridDim.x); if (ty == 0) isLastBlockDone = (value == (gridDim.x - 1)); } __syncthreads(); if(isLastBlockDone) { if(tx < gridDim.x) { *myTemp = partialSum[tx * blockDim.y + ty]; } __syncthreads(); for(int k = gridDim.x/2; k > 0; k/=2) { if(tx < k) { *myTemp +=*(myTemp + k); __syncthreads(); } } if(tx == 0) { counts[ty] = *myTemp; counter[ty] = 0; } } #ifdef USE_ARRAY_OF_ARRAYS if(tx == 0) free(temp[ty]); __syncthreads(); #endif if(tx == 0 && ty == 0) { #ifdef USE_SHM_MALLOC free(temp); #endif } }
Function: e_step
Wrapper-Function for the Kernel. Sets the threads distribution and runs them.
- Parameters:
-
cluster_assignment : array containing the cluster each data point belongs to. counts : array containing number of data points belonging to each cluster. data_points : the initial data point Matrix. cluster_centers : initial and later the calculated means of the clusters. n_clusters : number of clusters. n_pca : number of dimensions of a data point. n_data_points : number of data points.
- Returns:
- n_chg: number of changes in assignment of data points to cluster. If this Value is zero, then convergence is reached.
template<typename T> int _e_step(int * cluster_assignment, int * counts, const T * data_points, T * cluster_centers, const int n_clusters, const int n_pca, const int n_data_points) { int block_size = 512; int num_blocks = (n_data_points + block_size -1)/block_size; printf("\n%s BEGIN: num_blocks for e_step KERNEL: %d\n", __FUNCTION__, num_blocks); fflush(stdout); int * t_changes = new int [num_blocks]; for(int i = 0; i< num_blocks; i++) t_changes[i] = 0; int * t_changes_d; cudaMalloc(&t_changes_d, num_blocks*sizeof(int)); int * n_points_per_cluster_h = new int[n_clusters*num_blocks]; int * n_points_per_cluster; cudaMalloc(&n_points_per_cluster, num_blocks*n_clusters*sizeof(int)); int * counts_d; cudaMalloc(&counts_d, n_clusters*sizeof(int));
starts the __e_step Kernel with tha calculated number of blocks and the needed block size
__e_step<<<num_blocks, block_size>>>(cluster_assignment,
data_points,
cluster_centers,
t_changes_d,
n_clusters,
n_pca,
n_data_points,
n_points_per_cluster);
cudaThreadSynchronize();
cudaMemcpy(n_points_per_cluster_h, n_points_per_cluster,
num_blocks*n_clusters*sizeof(int), cudaMemcpyDeviceToHost);
for(int j = 0; j< n_clusters; j++) counts[j] = 0;
sum of the counts array (CPU)
for(int i = 0; i<num_blocks;i++) for(int j = 0; j< n_clusters; j++) counts[j] += n_points_per_cluster_h[n_clusters*i+j]; int warpSize = 32; int blockDim_x = ((256/n_clusters)/warpSize)*warpSize; if(blockDim_x > num_blocks) { blockDim_x = ((num_blocks + warpSize-1)/warpSize) * warpSize; } int num_blocks_sum_counts = (num_blocks + blockDim_x-1)/blockDim_x; int power_2_n_clusters = log(num_blocks_sum_counts)/log(2.); num_blocks_sum_counts = pow(2, power_2_n_clusters); printf("num_blocks_sum_counts :%d \n", num_blocks_sum_counts); int n_points_per_cluster_size = num_blocks * n_clusters; dim3 block_size_sum(blockDim_x, n_clusters); int * partialSum; cudaMalloc(&partialSum, n_clusters*num_blocks_sum_counts*sizeof(int)); unsigned int * counter; cudaMalloc(&counter, n_clusters*sizeof(unsigned int)); printf("__sum_counts_array : %d blocks of size (%d,%d)\n", num_blocks_sum_counts, blockDim_x, n_clusters); fflush(stdout); __sum_counts_array<<<num_blocks_sum_counts, block_size_sum>>>(n_points_per_cluster, n_points_per_cluster_size, counts_d, partialSum, counter); cudaThreadSynchronize(); int * counts_by_gpu = new int[n_clusters]; cudaMemcpy(counts_by_gpu, counts_d, n_clusters*sizeof(int), cudaMemcpyDeviceToHost); for(int i = 0; i < n_clusters; i++) { if (counts_by_gpu[i] != counts[i]) printf("CPU and GPU results differ! CPU : %d, GPU : %d\n", counts[i], counts_by_gpu[i]); else counts[i] = counts_by_gpu[i]; }
getting the array containing the number of changes in assignment back from the GPU
cudaMemcpy(t_changes, t_changes_d, num_blocks*sizeof(int), cudaMemcpyDeviceToHost);
sums the changes array to a single value
int n_changes = 0; for(int i=0; i<num_blocks; i++) n_changes += t_changes[i]; delete [] t_changes; delete [] counts_by_gpu; delete [] n_points_per_cluster_h; cudaFree(partialSum); cudaFree(counter); cudaFree(t_changes_d); cudaFree(n_points_per_cluster); cudaFree(counts_d); printf("n reassignments : %d\n", n_changes); return n_changes; }
Function : e_step
e_step wrapper for floats.
- Parameters:
-
cluster_assignment : array containing the cluster each data point belongs to. counts : array containing number of data points belonging to each cluster. data_points : the initial data point Matrix. cluster_centers : initial and later the calculated means of the clusters. n_clusters : number of clusters. n_pca : number of dimensions of a data point. n_data_points : number of data points.
- Returns:
- result: number of changes in assignment of data points to cluster.
int e_step(int * cluster_assignment, int * counts, const float * data_points, float * cluster_centers, const int n_clusters, const int n_pca, const int n_data_points) { int result = _e_step(cluster_assignment, counts, data_points, cluster_centers, n_clusters, n_pca, n_data_points); return result; } #ifndef USE_SINGLE_PRECISION
Function : e_step
e_step wrapper for double.
- Parameters:
-
cluster_assignment : array containing the cluster each data point belongs to. counts : array containing number of data points belonging to each cluster. data_points : the initial data point Matrix. cluster_centers : initial and later the calculated means of the clusters. n_clusters : number of clusters. n_pca : number of dimensions of a data point. n_data_points : number of data points.
- Returns:
- result: number of changes in assignment of data points to cluster.
int e_step(int * cluster_assignment, int * counts, const double * data_points, double * cluster_centers, const int n_clusters, const int n_pca, const int n_data_points) { int result = _e_step(cluster_assignment, counts, data_points, cluster_centers, n_clusters, n_pca, n_data_points); return result; } #endif #ifndef CUDADriver_STEP_6_H #define CUDADriver_STEP_6_H #include <lac/full_matrix.h> #include <lac/vector.h> #include <cmath> #include <step-1/cuda_driver_step-1.h> #include <step-1/cuda_driver_step-1.hh> #include <step-1/cuda_kernel_wrapper_step-1.cu.h> namespace step6 {
Class: Kmeans
Declares the methods and variables needed for the K-means algorithm
template<typename T, typename blas> class Kmeans { public:
blas++ Data types
typedef typename blas_pp<T, blas>::blas_wrapper_type BW; typedef typename blas_pp<T, blas>::FullMatrixAccessor FullMatrixAccessor; typedef typename blas_pp<T, blas>::Vector Vector; typedef typename blas_pp<int, blas>::Vector IntVector;
Kmeans(::FullMatrix<T> &ddata, ::FullMatrix<T> &mmeans, int openMP_threads);
Kmeans(FullMatrixAccessor &ddata, ::FullMatrix<T> &mmeans, int openMP_threads); int n_data_points, dim, n_clusters, n_threads; std::vector<int> n_chgs;
FullMatrix<T> data;
FullMatrixAccessor data;
::SmartPointer<::FullMatrix<T> > means;
std::vector<int> assign;
std::vector<std::vector<int> > counts;
int estep();
void mstep();
void fstep();
int estep_gpu();
void mstep_gpu();
void initial_computation(QString method);
void iterate(QString method,int max_iteration);
~Kmeans();
};
} // namespace step6 END
#endif // CUDADriver_STEP_6_H
#ifndef CUDA_DRIVER_STEP_6_HH
#define CUDA_DRIVER_STEP_6_HH
#include "cuda_driver_step-6.h"
Declaration of Kernel-Wrapper-Functions.
#include <step-6/cuda_kernel_wrapper_step-6.cu.h> #include <step-1/cuda_kernel_wrapper_step-1.cu.h> #include <cutil_inline.h> #include <numeric> #include <omp.h> #include <QTime> #include <iostream> #include <fstream>
Constructor: Kmeans
Initialize the Kmeans algorithm the initial data points and means are passed from the user input in the .prm file
- Parameters:
-
ddata : contains the test data points mmeans : contains the initial means at the start of the simulation, with every iteration of the algorithm new means are calculated. The algorithm consist of 2 Steps, Expectation E-step and Maximization M-Step
it goes as follows:
Minimize the Function:
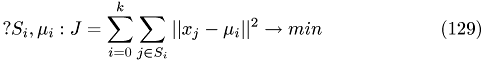
Initializing: set k random cluster centers, so-called means

Assign each data point to the nearest cluster center
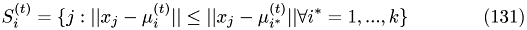
Update : recompute the cluster centers
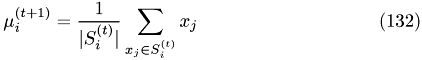
Convergence when :

template<typename T, typename blas> step6::Kmeans<T, blas>::Kmeans(/ *::FullMatrix<T>* / FullMatrixAccessor &ddata, ::FullMatrix<T> &mmeans, int openMP_threads) : n_data_points(ddata.n_rows()), dim(ddata.n_cols()), n_threads(openMP_threads), n_clusters(mmeans.n_rows()), data(ddata), means(&mmeans), assign(n_data_points), counts(n_threads, std::vector<int> (n_clusters)), n_chgs(n_threads) { BW::Init(); }
Function: initial_computation
This function calls the fstep(), mstep() and estep() once to creates the initial data for the Kmeans++ algorithm
- Parameters:
-
method : user decides, whether to use the CPU or the GPU method
template<typename T, typename blas> void step6::Kmeans<T, blas>::initial_computation(QString method) { if(QString::compare(method, "cpu", Qt::CaseInsensitive)==0){ QTime time; time.start(); fstep(); std::cout << "fstep cpu Time =" << time.elapsed() << std::endl; time.restart(); estep(); std::cout << "estep cpu Time =" << time.elapsed() << std::endl; time.restart(); mstep(); std::cout << "mstep cpu Time =" << time.elapsed() << std::endl; }else{ if(QString::compare(method, "gpu", Qt::CaseInsensitive)==0){ QTime time; time.start(); fstep(); std::cout << "fstep gpu Time =" << time.elapsed() << std::endl; time.restart(); estep_gpu(); std::cout << "estep gpu Time =" << time.elapsed() << std::endl; time.restart(); mstep(); std::cout << "mstep gpu Time =" << time.elapsed() << std::endl; } } }
Function: iterate
This function iterates over estep() and mstep() until convergence is reached (or max_iteration is reached) when the maximal iteration is reached, the means would be found in the means array.
- Parameters:
-
method : CPU or GPU method, decided by user max_iter : maximal number of iterations ( not used at this time )
template<typename T, typename blas> void step6::Kmeans<T, blas>::iterate(QString method, int max_iter) { if(QString::compare(method, "cpu", Qt::CaseInsensitive)==0){ QTime iTime, estepTime; iTime.start(); int n_changes = -1, n_changes_old = -2, iter = 0; estepTime.start(); n_changes_old = estep(); std::cout << "cpu estepTime =" << estepTime.elapsed() << std::endl; mstep(); while(n_changes != n_changes_old && iter <= max_iter) { n_changes_old = n_changes; n_changes = estep(); mstep(); iter++; } std::cout << "iterations cpu = "<< iter << std::endl; std::cout << "cpu iter Time =" << iTime.elapsed() << std::endl; }else{ if(QString::compare(method, "gpu", Qt::CaseInsensitive)==0){ QTime iTime, estepTime; iTime.start(); int n_changes = -1, n_changes_old = -2, iter = 0; estepTime.start(); n_changes_old = estep_gpu(); std::cout << "gpu estepTime =" << estepTime.elapsed() << std::endl; mstep(); while(n_changes != n_changes_old && iter <= max_iter) { n_changes_old = n_changes; n_changes = estep_gpu(); mstep(); iter++; } std::cout << "iterations gpu = "<< iter << std::endl; std::cout << "gpu iter Time =" << iTime.elapsed() << std::endl; } } }
Function: fstep
This function creates the initial means for the Kmeans++ algorithm before starting the actual Kmeans.
Kmeans++ is an algorithm for choosing the initial values for the Kmeans clustering.
the name Kmeans++ was given to this algorithm by David Arthur and Sergei Vassilvitskii in 2007 (Stanford University)
The algorithm is as follows:
1. Choose one center uniformly at random from among the data points.
2. For each data point x, compute 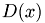 , the distance between x and the nearest center that has already been chosen.
, the distance between x and the nearest center that has already been chosen.
3. Add one new data point at random as a new center, using a weighted probability distribution where a point x is chosen with probability proportional to 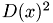 .
.
4. Repeat Steps 2 and 3 until k centers have been chosen.
5. Now that the initial centers have been chosen, proceed using standard Kmeans clustering.
(from Wikipedia, the free encyclopedia)
Implementation based on the implementation from Marius Muja (mariusm@cs.ubc.ca). All rights reserved. David G. Lowe (lowe@cs.ubc.ca). All rights reserved. copyright 2008-2009
template<typename T, typename blas> void step6::Kmeans<T, blas>::fstep() { omp_set_num_threads(4); double current_variance = 0; double result; std::vector<T> closest_dist_sq(n_data_points);
Choose one random center and set the closest_dist_sq values
int index = (int)(double(rand()) / RAND_MAX * n_data_points); for(int i = 0; i < dim; i++) (*means)(0, i) = data(index, i); result = 0; #pragma omp parallel for for (int i = 0; i < n_data_points; i++) { for(int j = 0; j < dim; j++){ result += ((data(i,j) - data(index,j)) * (data(i,j) - data(index,j))); } closest_dist_sq[i] = result; current_variance += closest_dist_sq[i]; }
Choose each center
for (int n_centers = 1; n_centers < n_clusters; n_centers++) {
Repeat several trials
double best_new_variance = -1; int best_new_index; for (int local_trial = 0; local_trial < 10; local_trial++) {
Choose our center - have to be slightly careful to return a valid answer even accounting for possible rounding errors
double rand_val = double(rand()) / RAND_MAX * current_variance; for (index = 0; index < n_data_points-1; index++) { if (rand_val <= closest_dist_sq[index]) break; else rand_val -= closest_dist_sq[index]; }
Compute the new variance
double new_variance = 0; result = 0; for (int i = 0; i < n_data_points; i++){ for(int j = 0; j < dim; j++) result += ((data(i,j) - data(index,j)) * (data(i,j) - data(index,j))); new_variance += std::min( result, closest_dist_sq[i] ); }
Store the best result
if (best_new_variance < 0 || new_variance < best_new_variance) {
best_new_variance = new_variance;
best_new_index = index;
}
}
Add the appropriate center
for(int i = 0; i < dim; i++) (*means)(n_centers, i) = data(best_new_index, i); current_variance = best_new_variance; result = 0; #pragma omp parallel for for (int i = 0; i < n_data_points; i++){ for(int j = 0; j < dim; j++) result += ((data(i,j) - data(best_new_index,j)) * (data(i,j) - data(best_new_index,j))); closest_dist_sq[i] = std::min( result, closest_dist_sq[i] ); } } }
Function: estep
The E-step of the Kmeans algorithm based on the Lloyd's algorithm.
The E-step calculates the squared distances between the data points and the means and assigns every data point to one cluster mean.
It also calculates the counts array containing the number of data points belonging to each cluster.
- Returns:
- n_chg : number of changes in data point to cluster assignment, if this number is zero, the algorithm has reached convergence and should terminate.
template<typename T, typename blas> int step6::Kmeans<T, blas>::estep() { int n_dp_per_thread = n_data_points / n_threads; if((n_data_points%n_threads)!= 0) n_dp_per_thread++; omp_set_num_threads(n_threads); #pragma omp parallel for for(int thread=0; thread<n_threads; thread++){ n_chgs[thread] = 0; for(int c = 0; c < counts[thread].size(); c++) counts[thread][c]=0; int dp_begin = thread*n_dp_per_thread; int dp_end = dp_begin+n_dp_per_thread; if(n_data_points - dp_begin < n_dp_per_thread) dp_end = dp_begin +(n_data_points - dp_begin); int kmin; double dmin; for (int n=dp_begin;n<dp_end;n++){ dmin = 9.99e99; for (int k=0;k<n_clusters;k++) { double d = 0.; for (int m=0; m<dim; m++) d+= std::pow(data(n,m) - (*means)(k,m), 2); if (d < dmin){ dmin = d; kmin = k; } } if (kmin != assign[n]) n_chgs[thread]++; assign[n] = kmin; counts[thread][kmin]++; } }
sums up the counts array from each thread (openMP)
for(int thread=1; thread<n_threads; thread++){ for(int i=0; i<counts[thread].size(); i++) counts[0][i]+= counts[thread][i]; }
sums up the number of changes in assignments
int n_chg = std::accumulate(n_chgs.begin(), n_chgs.end(), 0); return n_chg; }
Function: mstep
The M-step of the Kmeans algorithm calculates the new means for each cluster by using the assign and counts arrays calculated in the E-step.
template<typename T, typename blas> void step6::Kmeans<T, blas>::mstep() { int n, k, m; *means = 0.; for (n=0;n<n_data_points;n++) for( m=0; m<dim; m++) (*means)(assign[n],m) += data(n,m); for (k=0;k<n_clusters;k++){ if (counts[0][k] > 0) for (m=0;m<dim;m++) (*means)(k,m)/= counts[0][k]; } }
Function: estep_gpu
GPU implementation of the E-step of the Kmeans algorithm This function calls the CUDA-Wrapper function e_step
- Returns:
- n_chg : number of changes in data point to cluster assignment, if this number is zero, the algorithm has reached convergence and should terminate.
template<typename T, typename blas> int step6::Kmeans<T, blas>::estep_gpu() { int n_chg = 0; FullMatrixAccessor data_h(data); std::vector<T> data_points_h(data_h.n_elements()); std::copy(data_h.val(),data_h.val()+data_h.n_elements(),data_points_h.begin()); Vector data_points_d; data_points_d = data_points_h; FullMatrixAccessor means_h(*means); std::vector<T> means_points_h(means_h.n_elements()); std::copy(means_h.val(), means_h.val()+means_h.n_elements(), means_points_h.begin()); Vector means_points_d; means_points_d = means_points_h; IntVector assign_d; assign_d = assign; n_chg = e_step(assign_d.array().val(), &counts[0][0], data_points_d.array().val(), means_points_d.array().val(), n_clusters, dim, n_data_points); cudaMemcpy(&assign[0], assign_d.array().val(), assign_d.size()*sizeof(int), cudaMemcpyDeviceToHost); return n_chg; }
Destructor : ~Kmeans
Free the memory taken by Kmeans and shuts down the blas library.
template<typename T, typename blas> step6::Kmeans<T, blas>::~Kmeans() { BW::Shutdown(); } #endif // CUDA_DRIVER_STEP_6_HH
Results
Results are from the hybrid version of Kmeans using the e_step GPU Kernel and the m_step on the CPU. Using randomly generated data points to the cluster means {(0,0) (0,4) (5,5) (8,0) (12,12) (15,15)}
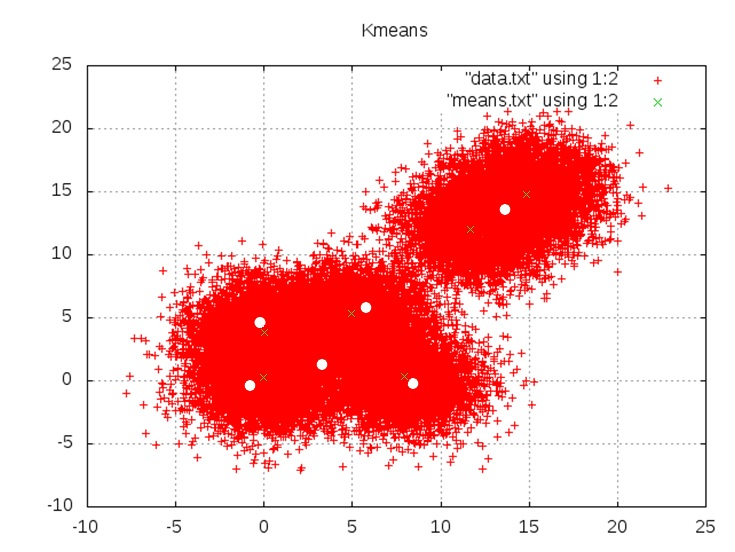
Kmeans
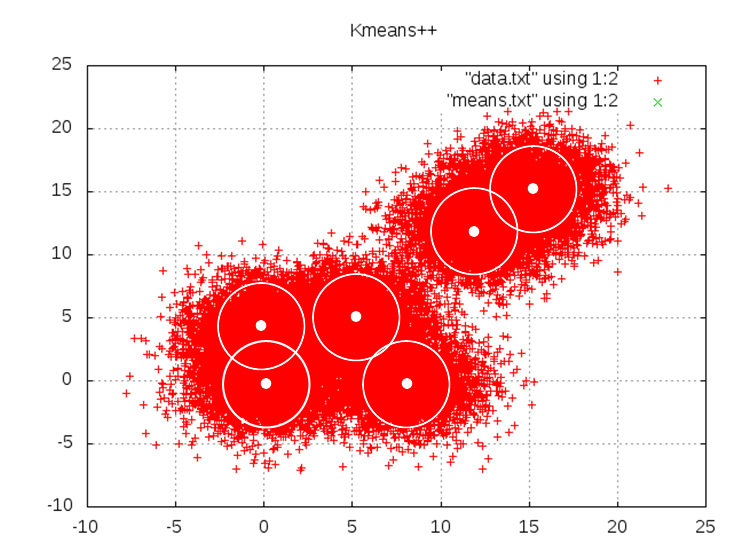
Kmeans++
K-means++ is an algorithm for choosing the initial values for the Kmeans clustering. the name Kmeans++ was given to this algorithm by David Arthur and Sergei Vassilvitskii in 2007 (Stanford University). The circles in the figure above represent the deviation the points are distributed with.
By choosing different starting points for the algorithm we can observe, that the error always converges to nearly zero and the algorithm find almost the same means every time.
(0,0) denotes the that every starting point is at (0,0), Diag being diagonal starting points as (0,0),(1,1),(2,2),... and Near are starting points near the actuall means with 0.2 variation.
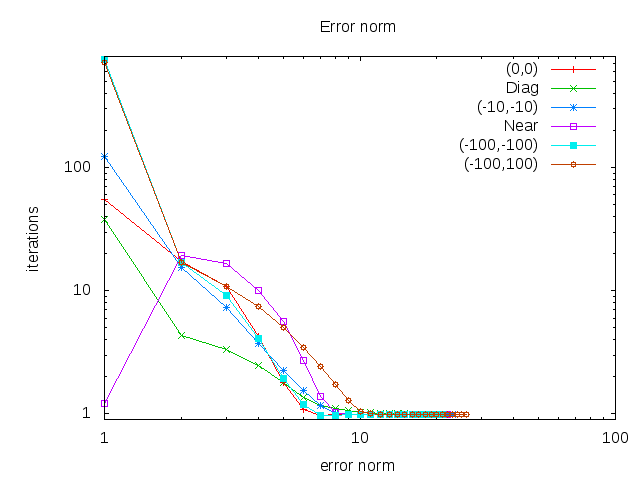
Error norm
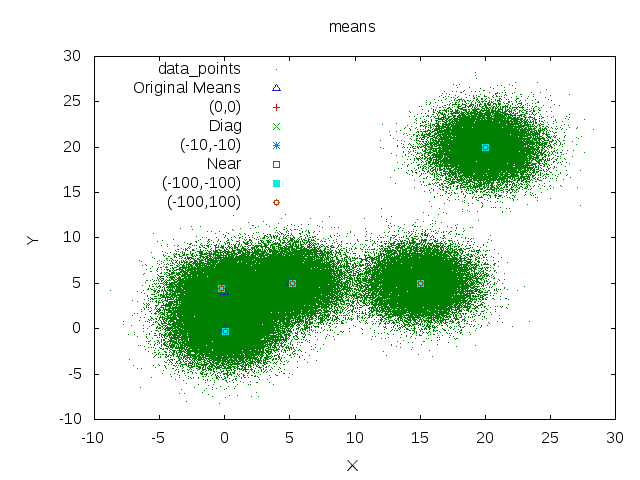
Means
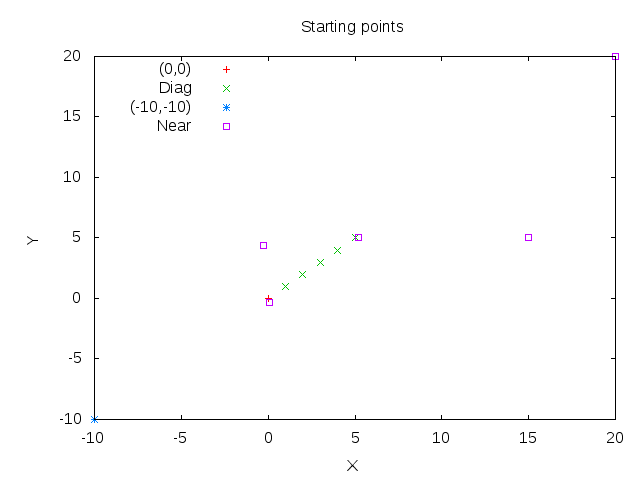
Starting points
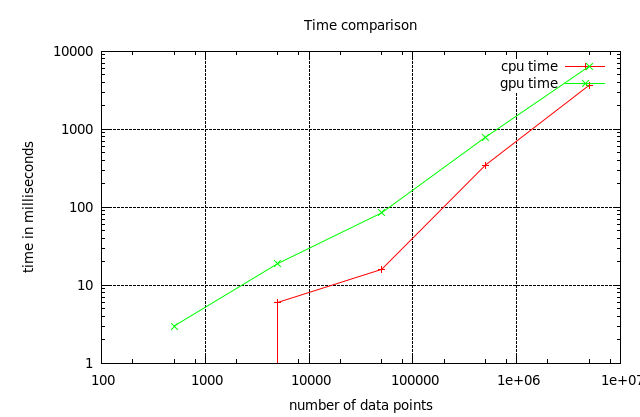
Times Comparison
Therefore using the CPU implementation is more efficient.
For the interested reader: computation of deceleration factor is left as an exercise.
The plain program
(If you are looking at a locally installed CUDA HPC Praktikum version, then the program can be found at .. /.. /testsite / /step-6 /step-cu.cc . Otherwise, this is only the path on some remote server.)
#include <iostream> #include <vector> #include <fstream> #include <QDir> #include <QString> #include <QVector> #include "Kmeans.h"
int main(int argc, char *argv[]) { cudaGetDeviceCount(&global_data.n_CUDA_devices); std::cout << "N available CUDA devices : " << global_data.n_CUDA_devices << std::endl; } #ifndef CUDA_KERNEL_STEP_6_CU_H #define CUDA_KERNEL_STEP_6_CU_H
int e_step(int * cluster_assignment, int * counts, const float * data_points, float * cluster_centers, const int n_clusters, const int n_pca, const int n_data_points); #ifndef USE_SINGLE_PRECISION int e_step(int * cluster_assignment, int * counts, const double * data_points, double * cluster_centers, const int n_clusters, const int n_pca, const int n_data_points); #endif #endif // CUDA_KERNEL_STEP_6_CU_H #include <cutil_inline.h> #include <step-6/cuda_kernel_wrapper_step-6.cu.h>
template <typename T> __global__ void __e_step(int * cluster_assignment, const T * data_points, const T * cluster_centers, int * t_changes, const int n_clusters, const int n_pca, const int n_data_points, int * n_points_per_cluster) { int warp_id = threadIdx.x/ WARP_SIZE; int n_warps = (blockDim.x +WARP_SIZE -1)/ WARP_SIZE; int i = threadIdx.x + blockDim.x * blockIdx.x; __shared__ int * n_changes; if (threadIdx.x == 0) n_changes = (int*) malloc (WARP_SIZE*sizeof(int)); __shared__ int * counts; if (threadIdx.x == 0) counts = (int*) malloc (n_clusters*sizeof(int)); __syncthreads(); if(threadIdx.x < WARP_SIZE) n_changes[threadIdx.x] = 0; if(threadIdx.x < n_clusters) counts[threadIdx.x] = 0; __syncthreads(); if(i>=n_data_points) return; int c_min = 0, c_min_old = -1; T d_min = 0.; T tmp, distance_ij; for(int d = 0; d < n_pca; d++) { tmp = data_points[n_pca*i+d]- cluster_centers[d]; d_min += tmp*tmp; c_min_old = cluster_assignment[i]; } for(int c = 1; c < n_clusters; c++) { distance_ij = 0; for(int d = 0; d < n_pca; d++) { tmp = data_points[n_pca*i+d]- cluster_centers[n_pca*c+d]; distance_ij += tmp*tmp; } if(distance_ij < d_min) { d_min = distance_ij; c_min = c; } } cluster_assignment[i] = c_min; atomicAdd(counts+c_min, 1); n_changes[warp_id] = abs(any(c_min - c_min_old)); __syncthreads(); if(threadIdx.x < n_clusters) n_points_per_cluster[n_clusters*blockIdx.x+threadIdx.x] = counts[threadIdx.x]; if(threadIdx.x == 0) { t_changes[blockIdx.x] = 0; for(int k = 0; k < n_warps; k++) t_changes[blockIdx.x] += n_changes[k]; __threadfence_block(); free(n_changes); free(counts); } }
__global__ void __sum_counts_array(const int * n_points_per_cluster, const int n_data_points, int * counts, int * partialSum, unsigned int * counter) { const int ty = threadIdx.y; const int tx = threadIdx.x; __shared__ bool isLastBlockDone; if(tx == 0 && ty == 0) isLastBlockDone = false; __syncthreads(); #ifdef USE_ARRAY_OF_ARRAYS __shared__ int ** temp; if (tx == 0 && ty == 0) temp = (int**) malloc (blockDim.y*sizeof(int*)); __syncthreads(); if(tx == 0) temp[ty] = (int*) malloc (blockDim.x*sizeof(int)); __syncthreads(); temp[ty][threadIdx.x] = 0; __syncthreads(); volatile int * myTemp = temp [ty] + tx; #else #ifdef USE_SHM_MALLOC __shared__ int * temp; if (tx == 0 && ty == 0) temp = (int*) malloc (blockDim.x*blockDim.y*sizeof(int)); #else __shared__ int temp[256 / *fixed size. thread blocks must not be larger! * / ]; #endif __syncthreads(); volatile int * myTemp = temp + blockDim.x*ty + tx; *myTemp = 0; __syncthreads(); #endif int global_index = ty + blockDim.y * tx + (blockDim.x*blockDim.y*blockIdx.x); if(global_index >= n_data_points) return; * myTemp = n_points_per_cluster[global_index]; __syncthreads(); for(int i = global_index + gridDim.x * blockDim.x*blockDim.y; i < n_data_points; i+= gridDim.x* blockDim.x*blockDim.y) { *myTemp += n_points_per_cluster[i]; __syncthreads(); } for(int k = blockDim.x/2; k > 0; k/=2) { if(tx < k) { *myTemp += *(myTemp + k); __syncthreads(); } } if(tx == 0) { partialSum[blockIdx.x * blockDim.y + ty] = *myTemp; __threadfence(); unsigned int value = atomicInc(&counter[ty], gridDim.x); if (ty == 0) isLastBlockDone = (value == (gridDim.x - 1)); } __syncthreads(); if(isLastBlockDone) { if(tx < gridDim.x) { *myTemp = partialSum[tx * blockDim.y + ty]; } __syncthreads(); for(int k = gridDim.x/2; k > 0; k/=2) { if(tx < k) { *myTemp +=*(myTemp + k); __syncthreads(); } } if(tx == 0) { counts[ty] = *myTemp; counter[ty] = 0; } } #ifdef USE_ARRAY_OF_ARRAYS if(tx == 0) free(temp[ty]); __syncthreads(); #endif if(tx == 0 && ty == 0) { #ifdef USE_SHM_MALLOC free(temp); #endif } }
template<typename T> int _e_step(int * cluster_assignment, int * counts, const T * data_points, T * cluster_centers, const int n_clusters, const int n_pca, const int n_data_points) { int block_size = 512; int num_blocks = (n_data_points + block_size -1)/block_size; printf("\n%s BEGIN: num_blocks for e_step KERNEL: %d\n", __FUNCTION__, num_blocks); fflush(stdout); int * t_changes = new int [num_blocks]; for(int i = 0; i< num_blocks; i++) t_changes[i] = 0; int * t_changes_d; cudaMalloc(&t_changes_d, num_blocks*sizeof(int)); int * n_points_per_cluster_h = new int[n_clusters*num_blocks]; int * n_points_per_cluster; cudaMalloc(&n_points_per_cluster, num_blocks*n_clusters*sizeof(int)); int * counts_d; cudaMalloc(&counts_d, n_clusters*sizeof(int)); __e_step<<<num_blocks, block_size>>>(cluster_assignment, data_points, cluster_centers, t_changes_d, n_clusters, n_pca, n_data_points, n_points_per_cluster); cudaThreadSynchronize(); cudaMemcpy(n_points_per_cluster_h, n_points_per_cluster, num_blocks*n_clusters*sizeof(int), cudaMemcpyDeviceToHost); for(int j = 0; j< n_clusters; j++) counts[j] = 0; for(int i = 0; i<num_blocks;i++) for(int j = 0; j< n_clusters; j++) counts[j] += n_points_per_cluster_h[n_clusters*i+j]; int warpSize = 32; int blockDim_x = ((256/n_clusters)/warpSize)*warpSize; if(blockDim_x > num_blocks) { blockDim_x = ((num_blocks + warpSize-1)/warpSize) * warpSize; } int num_blocks_sum_counts = (num_blocks + blockDim_x-1)/blockDim_x; int power_2_n_clusters = log(num_blocks_sum_counts)/log(2.); num_blocks_sum_counts = pow(2, power_2_n_clusters); printf("num_blocks_sum_counts :%d \n", num_blocks_sum_counts); int n_points_per_cluster_size = num_blocks * n_clusters; dim3 block_size_sum(blockDim_x, n_clusters); int * partialSum; cudaMalloc(&partialSum, n_clusters*num_blocks_sum_counts*sizeof(int)); unsigned int * counter; cudaMalloc(&counter, n_clusters*sizeof(unsigned int)); printf("__sum_counts_array : %d blocks of size (%d,%d)\n", num_blocks_sum_counts, blockDim_x, n_clusters); fflush(stdout); __sum_counts_array<<<num_blocks_sum_counts, block_size_sum>>>(n_points_per_cluster, n_points_per_cluster_size, counts_d, partialSum, counter); cudaThreadSynchronize(); int * counts_by_gpu = new int[n_clusters]; cudaMemcpy(counts_by_gpu, counts_d, n_clusters*sizeof(int), cudaMemcpyDeviceToHost); for(int i = 0; i < n_clusters; i++) { if (counts_by_gpu[i] != counts[i]) printf("CPU and GPU results differ! CPU : %d, GPU : %d\n", counts[i], counts_by_gpu[i]); else counts[i] = counts_by_gpu[i]; } cudaMemcpy(t_changes, t_changes_d, num_blocks*sizeof(int), cudaMemcpyDeviceToHost); int n_changes = 0; for(int i=0; i<num_blocks; i++) n_changes += t_changes[i]; delete [] t_changes; delete [] counts_by_gpu; delete [] n_points_per_cluster_h; cudaFree(partialSum); cudaFree(counter); cudaFree(t_changes_d); cudaFree(n_points_per_cluster); cudaFree(counts_d); printf("n reassignments : %d\n", n_changes); return n_changes; }
int e_step(int * cluster_assignment, int * counts, const float * data_points, float * cluster_centers, const int n_clusters, const int n_pca, const int n_data_points) { int result = _e_step(cluster_assignment, counts, data_points, cluster_centers, n_clusters, n_pca, n_data_points); return result; } #ifndef USE_SINGLE_PRECISION
int e_step(int * cluster_assignment, int * counts, const double * data_points, double * cluster_centers, const int n_clusters, const int n_pca, const int n_data_points) { int result = _e_step(cluster_assignment, counts, data_points, cluster_centers, n_clusters, n_pca, n_data_points); return result; } #endif #ifndef CUDADriver_STEP_6_H #define CUDADriver_STEP_6_H #include <lac/full_matrix.h> #include <lac/vector.h> #include <cmath> #include <step-1/cuda_driver_step-1.h> #include <step-1/cuda_driver_step-1.hh> #include <step-1/cuda_kernel_wrapper_step-1.cu.h> namespace step6 {
template<typename T, typename blas> class Kmeans { public: typedef typename blas_pp<T, blas>::blas_wrapper_type BW; typedef typename blas_pp<T, blas>::FullMatrixAccessor FullMatrixAccessor; typedef typename blas_pp<T, blas>::Vector Vector; typedef typename blas_pp<int, blas>::Vector IntVector; Kmeans(FullMatrixAccessor &ddata, ::FullMatrix<T> &mmeans, int openMP_threads); int n_data_points, dim, n_clusters, n_threads; std::vector<int> n_chgs; FullMatrixAccessor data; ::SmartPointer<::FullMatrix<T> > means; std::vector<int> assign; std::vector<std::vector<int> > counts; int estep(); void mstep(); void fstep(); int estep_gpu(); void mstep_gpu(); void initial_computation(QString method); void iterate(QString method,int max_iteration); ~Kmeans(); }; } // namespace step6 END #endif // CUDADriver_STEP_6_H #ifndef CUDA_DRIVER_STEP_6_HH #define CUDA_DRIVER_STEP_6_HH #include "cuda_driver_step-6.h" #include <step-6/cuda_kernel_wrapper_step-6.cu.h> #include <step-1/cuda_kernel_wrapper_step-1.cu.h> #include <cutil_inline.h> #include <numeric> #include <omp.h> #include <QTime> #include <iostream> #include <fstream>
template<typename T, typename blas> step6::Kmeans<T, blas>::Kmeans(/ *::FullMatrix<T>* / FullMatrixAccessor &ddata, ::FullMatrix<T> &mmeans, int openMP_threads) : n_data_points(ddata.n_rows()), dim(ddata.n_cols()), n_threads(openMP_threads), n_clusters(mmeans.n_rows()), data(ddata), means(&mmeans), assign(n_data_points), counts(n_threads, std::vector<int> (n_clusters)), n_chgs(n_threads) { BW::Init(); }
template<typename T, typename blas> void step6::Kmeans<T, blas>::initial_computation(QString method) { if(QString::compare(method, "cpu", Qt::CaseInsensitive)==0){ QTime time; time.start(); fstep(); std::cout << "fstep cpu Time =" << time.elapsed() << std::endl; time.restart(); estep(); std::cout << "estep cpu Time =" << time.elapsed() << std::endl; time.restart(); mstep(); std::cout << "mstep cpu Time =" << time.elapsed() << std::endl; }else{ if(QString::compare(method, "gpu", Qt::CaseInsensitive)==0){ QTime time; time.start(); fstep(); std::cout << "fstep gpu Time =" << time.elapsed() << std::endl; time.restart(); estep_gpu(); std::cout << "estep gpu Time =" << time.elapsed() << std::endl; time.restart(); mstep(); std::cout << "mstep gpu Time =" << time.elapsed() << std::endl; } } }
template<typename T, typename blas> void step6::Kmeans<T, blas>::iterate(QString method, int max_iter) { if(QString::compare(method, "cpu", Qt::CaseInsensitive)==0){ QTime iTime, estepTime; iTime.start(); int n_changes = -1, n_changes_old = -2, iter = 0; estepTime.start(); n_changes_old = estep(); std::cout << "cpu estepTime =" << estepTime.elapsed() << std::endl; mstep(); while(n_changes != n_changes_old && iter <= max_iter) { n_changes_old = n_changes; n_changes = estep(); mstep(); iter++; } std::cout << "iterations cpu = "<< iter << std::endl; std::cout << "cpu iter Time =" << iTime.elapsed() << std::endl; }else{ if(QString::compare(method, "gpu", Qt::CaseInsensitive)==0){ QTime iTime, estepTime; iTime.start(); int n_changes = -1, n_changes_old = -2, iter = 0; estepTime.start(); n_changes_old = estep_gpu(); std::cout << "gpu estepTime =" << estepTime.elapsed() << std::endl; mstep(); while(n_changes != n_changes_old && iter <= max_iter) { n_changes_old = n_changes; n_changes = estep_gpu(); mstep(); iter++; } std::cout << "iterations gpu = "<< iter << std::endl; std::cout << "gpu iter Time =" << iTime.elapsed() << std::endl; } } }
template<typename T, typename blas> void step6::Kmeans<T, blas>::fstep() { omp_set_num_threads(4); double current_variance = 0; double result; std::vector<T> closest_dist_sq(n_data_points); int index = (int)(double(rand()) / RAND_MAX * n_data_points); for(int i = 0; i < dim; i++) (*means)(0, i) = data(index, i); result = 0; #pragma omp parallel for for (int i = 0; i < n_data_points; i++) { for(int j = 0; j < dim; j++){ result += ((data(i,j) - data(index,j)) * (data(i,j) - data(index,j))); } closest_dist_sq[i] = result; current_variance += closest_dist_sq[i]; } for (int n_centers = 1; n_centers < n_clusters; n_centers++) { double best_new_variance = -1; int best_new_index; for (int local_trial = 0; local_trial < 10; local_trial++) { double rand_val = double(rand()) / RAND_MAX * current_variance; for (index = 0; index < n_data_points-1; index++) { if (rand_val <= closest_dist_sq[index]) break; else rand_val -= closest_dist_sq[index]; } double new_variance = 0; result = 0; for (int i = 0; i < n_data_points; i++){ for(int j = 0; j < dim; j++) result += ((data(i,j) - data(index,j)) * (data(i,j) - data(index,j))); new_variance += std::min( result, closest_dist_sq[i] ); } if (best_new_variance < 0 || new_variance < best_new_variance) { best_new_variance = new_variance; best_new_index = index; } } for(int i = 0; i < dim; i++) (*means)(n_centers, i) = data(best_new_index, i); current_variance = best_new_variance; result = 0; #pragma omp parallel for for (int i = 0; i < n_data_points; i++){ for(int j = 0; j < dim; j++) result += ((data(i,j) - data(best_new_index,j)) * (data(i,j) - data(best_new_index,j))); closest_dist_sq[i] = std::min( result, closest_dist_sq[i] ); } } }
template<typename T, typename blas> int step6::Kmeans<T, blas>::estep() { int n_dp_per_thread = n_data_points / n_threads; if((n_data_points%n_threads)!= 0) n_dp_per_thread++; omp_set_num_threads(n_threads); #pragma omp parallel for for(int thread=0; thread<n_threads; thread++){ n_chgs[thread] = 0; for(int c = 0; c < counts[thread].size(); c++) counts[thread][c]=0; int dp_begin = thread*n_dp_per_thread; int dp_end = dp_begin+n_dp_per_thread; if(n_data_points - dp_begin < n_dp_per_thread) dp_end = dp_begin +(n_data_points - dp_begin); int kmin; double dmin; for (int n=dp_begin;n<dp_end;n++){ dmin = 9.99e99; for (int k=0;k<n_clusters;k++) { double d = 0.; for (int m=0; m<dim; m++) d+= std::pow(data(n,m) - (*means)(k,m), 2); if (d < dmin){ dmin = d; kmin = k; } } if (kmin != assign[n]) n_chgs[thread]++; assign[n] = kmin; counts[thread][kmin]++; } } for(int thread=1; thread<n_threads; thread++){ for(int i=0; i<counts[thread].size(); i++) counts[0][i]+= counts[thread][i]; } int n_chg = std::accumulate(n_chgs.begin(), n_chgs.end(), 0); return n_chg; }
template<typename T, typename blas> void step6::Kmeans<T, blas>::mstep() { int n, k, m; *means = 0.; for (n=0;n<n_data_points;n++) for( m=0; m<dim; m++) (*means)(assign[n],m) += data(n,m); for (k=0;k<n_clusters;k++){ if (counts[0][k] > 0) for (m=0;m<dim;m++) (*means)(k,m)/= counts[0][k]; } }
template<typename T, typename blas> int step6::Kmeans<T, blas>::estep_gpu() { int n_chg = 0; FullMatrixAccessor data_h(data); std::vector<T> data_points_h(data_h.n_elements()); std::copy(data_h.val(),data_h.val()+data_h.n_elements(),data_points_h.begin()); Vector data_points_d; data_points_d = data_points_h; FullMatrixAccessor means_h(*means); std::vector<T> means_points_h(means_h.n_elements()); std::copy(means_h.val(), means_h.val()+means_h.n_elements(), means_points_h.begin()); Vector means_points_d; means_points_d = means_points_h; IntVector assign_d; assign_d = assign; n_chg = e_step(assign_d.array().val(), &counts[0][0], data_points_d.array().val(), means_points_d.array().val(), n_clusters, dim, n_data_points); cudaMemcpy(&assign[0], assign_d.array().val(), assign_d.size()*sizeof(int), cudaMemcpyDeviceToHost); return n_chg; }
template<typename T, typename blas> step6::Kmeans<T, blas>::~Kmeans() { BW::Shutdown(); } #endif // CUDA_DRIVER_STEP_6_HH
 documentation generated on Mon Jul 18 12:05:12 2011 by
doxygen
1.5.7
documentation generated on Mon Jul 18 12:05:12 2011 by
doxygen
1.5.7